Barriers to chimpanzee gene flow at the south-east edge of their distribution
- PMID: 37277946
- PMCID: PMC10421595
- DOI: 10.1111/mec.16986
Barriers to chimpanzee gene flow at the south-east edge of their distribution
Abstract
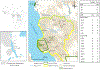
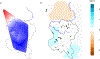
Populations on the edge of a species' distribution may represent an important source of adaptive diversity, yet these populations tend to be more fragmented and are more likely to be geographically isolated. Lack of genetic exchanges between such populations, due to barriers to animal movement, can not only compromise adaptive potential but also lead to the fixation of deleterious alleles. The south-eastern edge of chimpanzee distribution is particularly fragmented, and conflicting hypotheses have been proposed about population connectivity and viability. To address this uncertainty, we generated both mitochondrial and MiSeq-based microsatellite genotypes for 290 individuals ranging across western Tanzania. While shared mitochondrial haplotypes confirmed historical gene flow, our microsatellite analyses revealed two distinct clusters, suggesting two populations currently isolated from one another. However, we found evidence of high levels of gene flow maintained within each of these clusters, one of which covers an 18,000 km2 ecosystem. Landscape genetic analyses confirmed the presence of barriers to gene flow with rivers and bare habitats highly restricting chimpanzee movement. Our study demonstrates how advances in sequencing technologies, combined with the development of landscape genetics approaches, can resolve ambiguities in the genetic history of critical populations and better inform conservation efforts of endangered species.
Keywords: Tanzania; biogeography; genetic diversity; great apes; microsatellites; mitochondrial DNA.
© 2023 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Figures

References
-
- Alexander A (2015). genetic_diversity_diffs v1.0.0. Available from https://github.com/laninsky/genetic_diversity_diffs.
-
- Balkenhol N, Cushman S, Storfer A, & Waits L (2016). Landscape genetics: concepts, methods, applications.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

