Cell-Free Expression System Derived from a Near-Minimal Synthetic Bacterium
- PMID: 37278603
- PMCID: PMC10278164
- DOI: 10.1021/acssynbio.3c00114
Cell-Free Expression System Derived from a Near-Minimal Synthetic Bacterium
Abstract
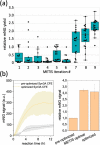
Cell-free expression (CFE) systems are fundamental to reconstituting metabolic pathways in vitro toward the construction of a synthetic cell. Although an Escherichia coli-based CFE system is well-established, simpler model organisms are necessary to understand the principles behind life-like behavior. Here, we report the successful creation of a CFE system derived from JCVI-syn3A (Syn3A), the minimal synthetic bacterium. Previously, high ribonuclease activity in Syn3A lysates impeded the establishment of functional CFE systems. Now, we describe how an unusual cell lysis method (nitrogen decompression) yielded Syn3A lysates with reduced ribonuclease activity that supported in vitro expression. To improve the protein yields in the Syn3A CFE system, we optimized the Syn3A CFE reaction mixture using an active machine learning tool. The optimized reaction mixture improved the CFE 3.2-fold compared to the preoptimized condition. This is the first report of a functional CFE system derived from a minimal synthetic bacterium, enabling further advances in bottom-up synthetic biology.
Keywords: JCVI-syn3A; Mycoplasma; active machine learning; cell-free expression system.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Gibson D. G.; Glass J. I.; Lartigue C.; Noskov V. N.; Chuang R.-Y.; Algire M. A.; Benders G. A.; Montague M. G.; Ma L.; Moodie M. M.; Merryman C.; Vashee S.; Krishnakumar R.; Assad-Garcia N.; Andrews-Pfannkoch C.; Denisova E. A.; Young L.; Qi Z.-Q.; Segall-Shapiro T. H.; Calvey C. H.; Parmar P. P.; Hutchison C. A.; Smith H. O.; Venter J. C. Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome. Science 2010, 329 (5987), 52–56. 10.1126/science.1190719. - DOI - PubMed
-
- Hutchison C. A.; Chuang R.-Y.; Noskov V. N.; Assad-Garcia N.; Deerinck T. J.; Ellisman M. H.; Gill J.; Kannan K.; Karas B. J.; Ma L.; Pelletier J. F.; Qi Z.-Q.; Richter R. A.; Strychalski E. A.; Sun L.; Suzuki Y.; Tsvetanova B.; Wise K. S.; Smith H. O.; Glass J. I.; Merryman C.; Gibson D. G.; Venter J. C. Design and Synthesis of a Minimal Bacterial Genome. Science 2016, 351 (6280), aad6253.10.1126/science.aad6253. - DOI - PubMed
-
- Breuer M.; Earnest T. M.; Merryman C.; Wise K. S.; Sun L.; Lynott M. R.; Hutchison C. A.; Smith H. O.; Lapek J. D.; Gonzalez D. J.; de Crécy-Lagard V.; Haas D.; Hanson A. D.; Labhsetwar P.; Glass J. I.; Luthey-Schulten Z.. Essential Metabolism for a Minimal Cell. Elife 2019, 8. 10.7554/eLife.36842. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

