SCCA1/SERPINB3 suppresses antitumor immunity and blunts therapy-induced T cell responses via STAT-dependent chemokine production
- PMID: 37279067
- PMCID: PMC10378164
- DOI: 10.1172/JCI163841
SCCA1/SERPINB3 suppresses antitumor immunity and blunts therapy-induced T cell responses via STAT-dependent chemokine production
Abstract
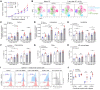
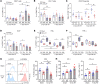
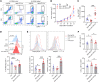
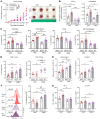
Patients with cancer who have high serum levels of squamous cell carcinoma antigen 1 (SCCA1, now referred to as SERPINB3) commonly experience treatment resistance and have a poor prognosis. Despite being a clinical biomarker, the modulation of SERPINB3 in tumor immunity is poorly understood. We found positive correlations of SERPINB3 with CXCL1, CXCL8 (CXCL8/9), S100A8, and S100A9 (S100A8/A9) myeloid cell infiltration through RNA-Seq analysis of human primary cervical tumors. Induction of SERPINB3 resulted in increased CXCL1/8 and S100A8/A9 expression, which promoted monocyte and myeloid-derived suppressor cell (MDSC) migration in vitro. In mouse models, Serpinb3a tumors showed increased MDSC and tumor-associated macrophage (TAM) infiltration, contributing to T cell inhibition, and this was further augmented upon radiation. Intratumoral knockdown (KD) of Serpinb3a resulted in tumor growth inhibition and reduced CXCL1 and S100A8/A expression and MDSC and M2 macrophage infiltration. These changes led to enhanced cytotoxic T cell function and sensitized tumors to radiotherapy (RT). We further revealed that SERPINB3 promoted STAT-dependent expression of chemokines, whereby inhibition of STAT activation by ruxolitinib or siRNA abrogated CXCL1/8 and S100A8/ A9 expression in SERPINB3 cells. Patients with elevated pretreatment SCCA levels and high phosphorylated STAT3 (p-STAT3) had increased intratumoral CD11b+ myeloid cells compared with patients with low SCCA levels and p-STAT3, who had improved overall survival after RT. These findings provide a preclinical rationale for targeting SERPINB3 in tumors to counteract immunosuppression and improve the response to RT.
Keywords: Cervical cancer; Oncology; Radiation therapy.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

