HvSL1 and HvMADS16 promote stamen identity to restrict multiple ovary formation in barley
- PMID: 37279531
- PMCID: PMC10498024
- DOI: 10.1093/jxb/erad218
HvSL1 and HvMADS16 promote stamen identity to restrict multiple ovary formation in barley
Abstract
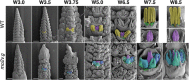
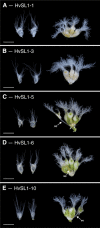
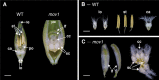
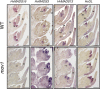
Correct floral development is the result of a sophisticated balance of molecular cues. Floral mutants provide insight into the main genetic determinants that integrate these cues, as well as providing opportunities to assess functional variation across species. In this study, we characterize the barley (Hordeum vulgare) multiovary mutants mov2.g and mov1, and propose causative gene sequences: a C2H2 zinc-finger gene HvSL1 and a B-class gene HvMADS16, respectively. In the absence of HvSL1, florets lack stamens but exhibit functional supernumerary carpels, resulting in multiple grains per floret. Deletion of HvMADS16 in mov1 causes homeotic conversion of lodicules and stamens into bract-like organs and carpels that contain non-functional ovules. Based on developmental, genetic, and molecular data, we propose a model by which stamen specification in barley is defined by HvSL1 acting upstream of HvMADS16. The present work identifies strong conservation of stamen formation pathways with other cereals, but also reveals intriguing species-specific differences. The findings lay the foundation for a better understanding of floral architecture in Triticeae, a key target for crop improvement.
Keywords: Triticeae; B-class; Barley; C2H2; MADS-box; cereal; flower development; multiovary; transcription factors.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ.. 2000. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Molecular Cell 5, 569–579. - PubMed
-
- Bai Y, Han N, Wu J, Yang Y, Wang J, Zhu M, Bian H.. 2014. A transient gene expression system using barley protoplasts to evaluate microRNAs for post-transcriptional regulation of their target genes. Plant Cell, Tissue and Organ Culture 119, 211–219.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

