Demographic history inference and the polyploid continuum
- PMID: 37279657
- PMCID: PMC10411560
- DOI: 10.1093/genetics/iyad107
Demographic history inference and the polyploid continuum
Abstract
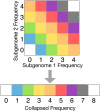
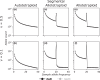
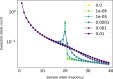
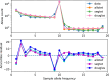
Polyploidy is an important generator of evolutionary novelty across diverse groups in the Tree of Life, including many crops. However, the impact of whole-genome duplication depends on the mode of formation: doubling within a single lineage (autopolyploidy) versus doubling after hybridization between two different lineages (allopolyploidy). Researchers have historically treated these two scenarios as completely separate cases based on patterns of chromosome pairing, but these cases represent ideals on a continuum of chromosomal interactions among duplicated genomes. Understanding the history of polyploid species thus demands quantitative inferences of demographic history and rates of exchange between subgenomes. To meet this need, we developed diffusion models for genetic variation in polyploids with subgenomes that cannot be bioinformatically separated and with potentially variable inheritance patterns, implementing them in the dadi software. We validated our models using forward SLiM simulations and found that our inference approach is able to accurately infer evolutionary parameters (timing, bottleneck size) involved with the formation of auto- and allotetraploids, as well as exchange rates in segmental allotetraploids. We then applied our models to empirical data for allotetraploid shepherd's purse (Capsella bursa-pastoris), finding evidence for allelic exchange between the subgenomes. Taken together, our model provides a foundation for demographic modeling in polyploids using diffusion equations, which will help increase our understanding of the impact of demography and selection in polyploid lineages.
Keywords: allopolyploidy; autopolyploidy; demography; homoeologous exchange; site frequency spectrum.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures
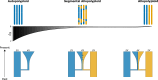


References
-
- Baduel P, Bray S, Vallejo-Marin M, Kolář F, Yant L. The “polyploid hop”: shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Front Ecol Evol. 2018;6:117. doi:10.3389/fevo.2018.00117 - DOI
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

