Colitis ameliorates cholestatic liver disease via suppression of bile acid synthesis
- PMID: 37280200
- PMCID: PMC10244448
- DOI: 10.1038/s41467-023-38840-8
Colitis ameliorates cholestatic liver disease via suppression of bile acid synthesis
Abstract
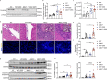
Primary sclerosing cholangitis (PSC) is a chronic cholestatic liver disease characterized by chronic inflammation and progressive fibrosis of the biliary tree. The majority of PSC patients suffer from concomitant inflammatory bowel disease (IBD), which has been suggested to promote disease development and progression. However, the molecular mechanisms by which intestinal inflammation may aggravate cholestatic liver disease remain incompletely understood. Here, we employ an IBD-PSC mouse model to investigate the impact of colitis on bile acid metabolism and cholestatic liver injury. Unexpectedly, intestinal inflammation and barrier impairment improve acute cholestatic liver injury and result in reduced liver fibrosis in a chronic colitis model. This phenotype is independent of colitis-induced alterations of microbial bile acid metabolism but mediated via hepatocellular NF-κB activation by lipopolysaccharide (LPS), which suppresses bile acid metabolism in-vitro and in-vivo. This study identifies a colitis-triggered protective circuit suppressing cholestatic liver disease and encourages multi-organ treatment strategies for PSC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

