Neoantigen-specific CD8 T cells with high structural avidity preferentially reside in and eliminate tumors
- PMID: 37280206
- PMCID: PMC10244384
- DOI: 10.1038/s41467-023-38946-z
Neoantigen-specific CD8 T cells with high structural avidity preferentially reside in and eliminate tumors
Abstract
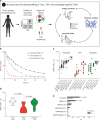
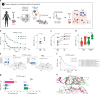
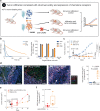
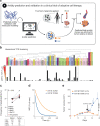
The success of cancer immunotherapy depends in part on the strength of antigen recognition by T cells. Here, we characterize the T cell receptor (TCR) functional (antigen sensitivity) and structural (monomeric pMHC-TCR off-rates) avidities of 371 CD8 T cell clones specific for neoantigens, tumor-associated antigens (TAAs) or viral antigens isolated from tumors or blood of patients and healthy donors. T cells from tumors exhibit stronger functional and structural avidity than their blood counterparts. Relative to TAA, neoantigen-specific T cells are of higher structural avidity and, consistently, are preferentially detected in tumors. Effective tumor infiltration in mice models is associated with high structural avidity and CXCR3 expression. Based on TCR biophysicochemical properties, we derive and apply an in silico model predicting TCR structural avidity and validate the enrichment in high avidity T cells in patients' tumors. These observations indicate a direct relationship between neoantigen recognition, T cell functionality and tumor infiltration. These results delineate a rational approach to identify potent T cells for personalized cancer immunotherapy.
© 2023. The Author(s).
Conflict of interest statement
G.C. has received grants, research support or is coinvestigator in clinical trials by Bristol-Myers-Squibb, Celgene, Boehringer Ingelheim, Tigen, Roche, Iovance and Kite. G.C. has received honoraria for consultations or presentations by Roche, Genentech, BMS, AstraZeneca, Sanofi-Aventis, Nextcure and GeneosTx. G.C. has patents in the domain of antibodies and vaccines targeting the tumor vasculature as well as technologies related to T cell expansion and engineering for T cell therapy. G.C. receives royalties from the University of Pennsylvania. S.B. and A.H. have patents in technologies related to T cell expansion and engineering for T cell therapy. Other authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

