EIF4A3-induced Circ_0001187 facilitates AML suppression through promoting ubiquitin-proteasomal degradation of METTL3 and decreasing m6A modification level mediated by miR-499a-5p/RNF113A pathway
- PMID: 37280654
- PMCID: PMC10243067
- DOI: 10.1186/s40364-023-00495-4
EIF4A3-induced Circ_0001187 facilitates AML suppression through promoting ubiquitin-proteasomal degradation of METTL3 and decreasing m6A modification level mediated by miR-499a-5p/RNF113A pathway
Abstract
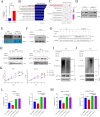
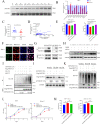
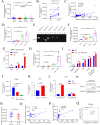
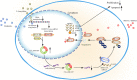
Aberrant expression of circRNAs has been proven to play a crucial role in the progression of acute myeloid leukemia (AML); however, its regulatory mechanism remains unclear. Herein, we identified a novel circRNA, Circ_0001187, which is downregulated in AML patients, and its low level contributes to a poor prognosis. We further validated their expression in large-scale samples and found that only the expression of Circ_0001187 was significantly decreased in newly diagnosed (ND) AML patients and increased in patients with hematological complete remission (HCR) compared with controls. Knockdown of Circ_0001187 significantly promoted proliferation and inhibited apoptosis of AML cells in vitro and in vivo, whereas overexpression of Circ _0001187 exerted the opposite effects. Interestingly, we found that Circ_0001187 decreases mRNA m6A modification in AML cells by enhancing METTL3 protein degradation. Mechanistically, Circ_0001187 sponges miR-499a-5p to enhance the expression of E3 ubiquitin ligase RNF113A, which mediates METTL3 ubiquitin/proteasome-dependent degradation via K48-linked polyubiquitin chains. Moreover, we found that the low expression of Circ _0001187 is regulated by promoter DNA methylation and histone acetylation. Collectively, our findings highlight the potential clinical implications of Circ _0001187 as a key tumor suppressor in AML via the miR-499a-5p/RNF113A/METTL3 pathway.
Keywords: Acute myeloid leukemia; Circ_0001187; METTL3; Progression; RNF113A; miR-499a-5p.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Circular RNA circ_0005774 contributes to proliferation and suppresses apoptosis of acute myeloid leukemia cells via circ_0005774/miR-192-5p/ULK1 ceRNA pathway.Biochem Biophys Res Commun. 2021 Apr 30;551:78-85. doi: 10.1016/j.bbrc.2021.02.058. Epub 2021 Mar 15. Biochem Biophys Res Commun. 2021. PMID: 33735626
-
Circ_0009910 sponges miR-491-5p to promote acute myeloid leukemia progression through modulating B4GALT5 expression and PI3K/AKT signaling pathway.Int J Lab Hematol. 2022 Apr;44(2):320-332. doi: 10.1111/ijlh.13742. Epub 2021 Oct 28. Int J Lab Hematol. 2022. PMID: 34709725
-
Tumor-suppressive MEG3 induces microRNA-493-5p expression to reduce arabinocytosine chemoresistance of acute myeloid leukemia cells by downregulating the METTL3/MYC axis.J Transl Med. 2022 Jun 27;20(1):288. doi: 10.1186/s12967-022-03456-x. J Transl Med. 2022. PMID: 35761379 Free PMC article.
-
Hsa_circ_0044907 promotes acute myeloid leukemia progression through upregulating oncogene KIT via sequestering miR-186-5p.Hematology. 2022 Dec;27(1):960-970. doi: 10.1080/16078454.2022.2113574. Hematology. 2022. PMID: 36004511
-
Long non-coding RNA LINC01018 inhibits the progression of acute myeloid leukemia by targeting miR-499a-5p to regulate PDCD4.Oncol Lett. 2021 Jul;22(1):541. doi: 10.3892/ol.2021.12802. Epub 2021 May 20. Oncol Lett. 2021. PMID: 34079594 Free PMC article.
Cited by
-
RNA m6A modification in ferroptosis: implications for advancing tumor immunotherapy.Mol Cancer. 2024 Sep 28;23(1):213. doi: 10.1186/s12943-024-02132-6. Mol Cancer. 2024. PMID: 39342168 Free PMC article. Review.
-
Small molecule inhibitors targeting m6A regulators.J Hematol Oncol. 2024 May 6;17(1):30. doi: 10.1186/s13045-024-01546-5. J Hematol Oncol. 2024. PMID: 38711100 Free PMC article. Review.
-
Recent Progress of CircRNAs in Hematological Malignancies.Int J Med Sci. 2024 Sep 30;21(13):2544-2561. doi: 10.7150/ijms.98156. eCollection 2024. Int J Med Sci. 2024. PMID: 39439468 Free PMC article. Review.
-
The crosstalk between alternative splicing and circular RNA in cancer: pathogenic insights and therapeutic implications.Cell Mol Biol Lett. 2024 Nov 16;29(1):142. doi: 10.1186/s11658-024-00662-x. Cell Mol Biol Lett. 2024. PMID: 39550559 Free PMC article. Review.
-
Circular RNA as Diagnostic and Prognostic Biomarkers in Hematological Malignancies:Systematic Review.Technol Cancer Res Treat. 2024 Jan-Dec;23:15330338241285149. doi: 10.1177/15330338241285149. Technol Cancer Res Treat. 2024. PMID: 39512224 Free PMC article.
References
LinkOut - more resources
Full Text Sources

