Suspension culture improves iPSC expansion and pluripotency phenotype
- PMID: 37280707
- PMCID: PMC10245469
- DOI: 10.1186/s13287-023-03382-9
Suspension culture improves iPSC expansion and pluripotency phenotype
Abstract
Background: Induced pluripotent stem cells (iPSCs) offer potential to revolutionize regenerative medicine as a renewable source for islets, dopaminergic neurons, retinal cells, and cardiomyocytes. However, translation of these regenerative cell therapies requires cost-efficient mass manufacturing of high-quality human iPSCs. This study presents an improved three-dimensional Vertical-Wheel® bioreactor (3D suspension) cell expansion protocol with comparison to a two-dimensional (2D planar) protocol.
Methods: Sendai virus transfection of human peripheral blood mononuclear cells was used to establish mycoplasma and virus free iPSC lines without common genetic duplications or deletions. iPSCs were then expanded under 2D planar and 3D suspension culture conditions. We comparatively evaluated cell expansion capacity, genetic integrity, pluripotency phenotype, and in vitro and in vivo pluripotency potential of iPSCs.
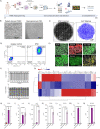
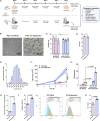
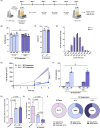
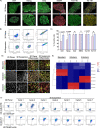
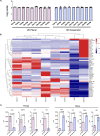
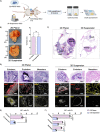
Results: Expansion of iPSCs using Vertical-Wheel® bioreactors achieved 93.8-fold (IQR 30.2) growth compared to 19.1 (IQR 4.0) in 2D (p < 0.0022), the largest expansion potential reported to date over 5 days. 0.5 L Vertical-Wheel® bioreactors achieved similar expansion and further reduced iPSC production cost. 3D suspension expanded cells had increased proliferation, measured as Ki67+ expression using flow cytometry (3D: 69.4% [IQR 5.5%] vs. 2D: 57.4% [IQR 10.9%], p = 0.0022), and had a higher frequency of pluripotency marker (Oct4+Nanog+Sox2+) expression (3D: 94.3 [IQR 1.4] vs. 2D: 52.5% [IQR 5.6], p = 0.0079). q-PCR genetic analysis demonstrated a lack of duplications or deletions at the 8 most commonly mutated regions within iPSC lines after long-term passaging (> 25). 2D-cultured cells displayed a primed pluripotency phenotype, which transitioned to naïve after 3D-culture. Both 2D and 3D cells were capable of trilineage differentiation and following teratoma, 2D-expanded cells generated predominantly solid teratomas, while 3D-expanded cells produced more mature and predominantly cystic teratomas with lower Ki67+ expression within teratomas (3D: 16.7% [IQR 3.2%] vs.. 2D: 45.3% [IQR 3.0%], p = 0.002) in keeping with a naïve phenotype.
Conclusion: This study demonstrates nearly 100-fold iPSC expansion over 5-days using our 3D suspension culture protocol in Vertical-Wheel® bioreactors, the largest cell growth reported to date. 3D expanded cells showed enhanced in vitro and in vivo pluripotency phenotype that may support more efficient scale-up strategies and safer clinical implementation.
Keywords: Bioreactor; Cell therapy; Expansion; Human-induced pluripotent stem cells; Pluripotency; Stem cells; iPSC.
© 2023. The Author(s).
Conflict of interest statement
AMJS serves as a consultant to ViaCyte Inc., Vertex Inc., Betalin Ltd., Hemostemix Inc. and Aspect Biosystems Ltd. No conflict of interest exists for all other co-authors.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

