An angiotensin converting enzyme homolog is required for volatile pheromone detection, odorant binding protein secretion and normal courtship behavior in Drosophila melanogaster
- PMID: 37283550
- PMCID: PMC10484059
- DOI: 10.1093/genetics/iyad109
An angiotensin converting enzyme homolog is required for volatile pheromone detection, odorant binding protein secretion and normal courtship behavior in Drosophila melanogaster
Abstract
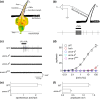
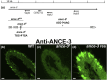
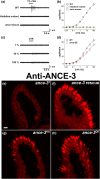
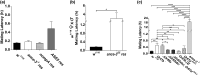
In many arthropods, including insects responsible for transmission of human diseases, behaviors that include mating, aggregation, and aggression are triggered by detection of pheromones. Extracellular odorant binding proteins are critical for pheromone detection in many insects and are secreted into the fluid bathing the olfactory neuron dendrites. In Drosophila melanogaster, the odorant binding protein LUSH is essential for normal sensitivity to the volatile sex pheromone, 11-cis vaccenyl acetate (cVA). Using a genetic screen for cVA pheromone insensitivity, we identified ANCE-3, a homolog of human angiotensin converting enzyme that is required for detection of cVA pheromone. The mutants have normal dose-response curves for food odors, although olfactory neuron amplitudes are reduced in all olfactory neurons examined. ance-3 mutants have profound delays in mating, and the courtship defects are primarily but not exclusively due to loss of ance-3 function in males. We demonstrate that ANCE-3 is required in the sensillae support cells for normal reproductive behavior, and that localization of odorant binding proteins to the sensillum lymph is blocked in the mutants. Expression of an ance-3 cDNA in sensillae support cells completely rescues the cVA responses, LUSH localization, and courtship defects. We show the courtship latency defects are not due to effects on olfactory neurons in the antenna nor mediated through ORCO receptors, but instead stem from ANCE-3-dependent effects on chemosensory sensillae in other body parts. These findings reveal an unexpected factor critical for pheromone detection with profound influence on reproductive behaviors.
Keywords: ACE; mating; olfaction; olfactory; reproduction.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

