Viable SARS-CoV-2 Omicron sub-variants isolated from autopsy tissues
- PMID: 37283920
- PMCID: PMC10240073
- DOI: 10.3389/fmicb.2023.1192832
Viable SARS-CoV-2 Omicron sub-variants isolated from autopsy tissues
Abstract
Introduction: Pulmonary and extrapulmonary manifestations have been described after infection with SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19). The virus is known to persist in multiple organs due to its tropism for several tissues. However, previous reports were unable to provide definitive information about whether the virus is viable and transmissible. It has been hypothesized that the persisting reservoirs of SARS-CoV-2 in tissues could be one of the multiple potentially overlapping causes of long COVID.
Methods: In the present study, we investigated autopsy materials obtained from 21 cadaveric donors with documented first infection or reinfection at the time of death. The cases studied included recipients of different formulations of COVID-19 vaccines. The aim was to find the presence of SARS-CoV-2 in the lungs, heart, liver, kidneys, and intestines. We used two technical approaches: the detection and quantification of viral genomic RNA using RT-qPCR, and virus infectivity using permissive in vitro Vero E6 culture.
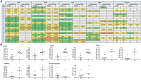
Results: All tissues analyzed showed the presence of SARS-CoV-2 genomic RNA but at dissimilar levels ranging from 1.01 × 102 copies/mL to 1.14 × 108 copies/mL, even among those cases who had been COVID-19 vaccinated. Importantly, different amounts of replication-competent virus were detected in the culture media from the studied tissues. The highest viral load were measured in the lung (≈1.4 × 106 copies/mL) and heart (≈1.9 × 106 copies/mL) samples. Additionally, based on partial Spike gene sequences, SARS-CoV-2 characterization revealed the presence of multiple Omicron sub-variants exhibiting a high level of nucleotide and amino acid identity among them.
Discussion: These findings highlight that SARS-CoV-2 can spread to multiple tissue locations such as the lungs, heart, liver, kidneys, and intestines, both after primary infection and after reinfections with the Omicron variant, contributing to extending knowledge about the pathogenesis of acute infection and understanding the sequelae of clinical manifestations that are observed during post-acute COVID-19.
Keywords: SARS-CoV-2; Vero E6 cell line; omicron variants; postmortem tissue; virus isolation.
Copyright © 2023 Maffia-Bizzozero, Cevallos, Remes Lenicov, Freiberger, Lopez, Guano Toaquiza, Sviercz, Jarmoluk, Bustos, D’Addario, Quarleri and Delpino.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

