COVID-19 impedimetric biosensor based on polypyrrole nanotubes, nickel hydroxide and VHH antibody fragment: specific, sensitive, and rapid viral detection in saliva samples
- PMID: 37284350
- PMCID: PMC10236006
- DOI: 10.1016/j.mtchem.2023.101597
COVID-19 impedimetric biosensor based on polypyrrole nanotubes, nickel hydroxide and VHH antibody fragment: specific, sensitive, and rapid viral detection in saliva samples
Abstract
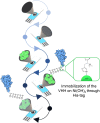
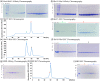
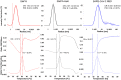
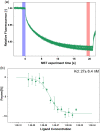
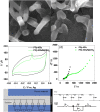
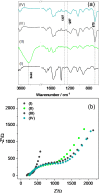
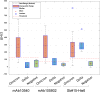
SARS-CoV-2 rapid spread required urgent, accurate, and prompt diagnosis to control the virus dissemination and pandemic management. Several sensors were developed using different biorecognition elements to obtain high specificity and sensitivity. However, the task to achieve these parameters in combination with fast detection, simplicity, and portability to identify the biorecognition element even in low concentration remains a challenge. Therefore, we developed an electrochemical biosensor based on polypyrrole nanotubes coupled via Ni(OH)2 ligation to an engineered antigen-binding fragment of heavy chain-only antibodies (VHH) termed Sb#15. Herein we report Sb#15-His6 expression, purification, and characterization of its interaction with the receptor-binding domain (RBD) of SARS-CoV-2 in addition to the construction and validation of a biosensor. The recombinant Sb#15 is correctly folded and interacts with the RBD with a dissociation constant (KD) of 27.1 ± 6.4 nmol/L. The biosensing platform was developed using polypyrrole nanotubes and Ni(OH)2, which can properly orientate the immobilization of Sb#15-His6 at the electrode surface through His-tag interaction for the sensitive SARS-CoV-2 antigen detection. The quantification limit was determined as 0.01 pg/mL using recombinant RBD, which was expressively lower than commercial monoclonal antibodies. In pre-characterized saliva, both Omicron and Delta SARS-CoV-2 were accurately detected only in positive samples, meeting all the requirements recommended by the World Health Organization for in vitro diagnostics. A low sample volume of saliva is needed to perform the detection, providing results within 15 min without further sample preparations. In summary, a new perspective allying recombinant VHHs with biosensor development and real sample detection was explored, addressing the need for accurate, rapid, and sensitive biosensors.
Keywords: COVID-19; Chemometrics; Impedimetric biosensor; Nanostructured electrode; Saliva sample.
© 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Gruell H., Vanshylla K., Korenkov M., Kurth F., Kreer C., Klein F. SARS-CoV-2 Omicron sublineages exhibit distinct antibody escape patterns short article SARS-CoV-2 Omicron sublineages exhibit distinct antibody escape patterns. Cell Host Microbe. 2022;30:1–11. doi: 10.1016/j.chom.2022.07.002. - DOI - PMC - PubMed
-
- Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Marschalek R., Herrmann E., Helfritz F.A., Wolf T., Goetsch U., Ciesek S. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies. EBioMedicine. 2022;82 doi: 10.1016/j.ebiom.2022.104158. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
