Spatial Transcriptomics of Intraductal Papillary Mucinous Neoplasms of the Pancreas Identifies NKX6-2 as a Driver of Gastric Differentiation and Indolent Biological Potential
- PMID: 37285225
- PMCID: PMC10880589
- DOI: 10.1158/2159-8290.CD-22-1200
Spatial Transcriptomics of Intraductal Papillary Mucinous Neoplasms of the Pancreas Identifies NKX6-2 as a Driver of Gastric Differentiation and Indolent Biological Potential
Abstract
Intraductal papillary mucinous neoplasms (IPMN) of the pancreas are bona fide precursor lesions of pancreatic ductal adenocarcinoma (PDAC). The most common subtype of IPMNs harbors a gastric foveolar-type epithelium, and these low-grade mucinous neoplasms are harbingers of IPMNs with high-grade dysplasia and cancer. The molecular underpinning of gastric differentiation in IPMNs is unknown, although identifying drivers of this indolent phenotype might enable opportunities for intercepting progression to high-grade IPMN and cancer. We conducted spatial transcriptomics on a cohort of IPMNs, followed by orthogonal and cross-species validation studies, which established the transcription factor NKX6-2 as a key determinant of gastric cell identity in low-grade IPMNs. Loss of NKX6-2 expression is a consistent feature of IPMN progression, while reexpression of Nkx6-2 in murine IPMN lines recapitulates the aforementioned gastric transcriptional program and glandular morphology. Our study identifies NKX6-2 as a previously unknown transcription factor driving indolent gastric differentiation in IPMN pathogenesis.
Significance: Identification of the molecular features driving IPMN development and differentiation is critical to prevent cancer progression and enhance risk stratification. We used spatial profiling to characterize the epithelium and microenvironment of IPMN, which revealed a previously unknown link between NKX6-2 and gastric differentiation, the latter associated with indolent biological potential. See related commentary by Ben-Shmuel and Scherz-Shouval, p. 1768. This article is highlighted in the In This Issue feature, p. 1749.
©2023 American Association for Cancer Research.
Conflict of interest statement
Conflict of Interest
A.M. receives royalties for a pancreatic cancer biomarker test from Cosmos Wisdom Biotechnology, and this financial relationship is managed and monitored by the UTMDACC Conflict of Interest Committee. A.M. is also listed as an inventor on a patent that has been licensed by Johns Hopkins University to ThriveEarlier Detection. A.M. serves as a consultant for Freenome and Tezcat Biotechnology. M.K. is a consultant for PanTher Therapeutics.
Figures

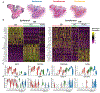
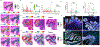



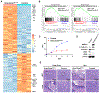
Comment in
-
Keeping IPMNs in Check: A Novel Role for the Transcription Factor NKX6-2 in Preserving an Indolent Cell Identity in Pancreatic Cystic Lesions.Cancer Discov. 2023 Aug 4;13(8):1768-1770. doi: 10.1158/2159-8290.CD-23-0590. Cancer Discov. 2023. PMID: 37539476
References
-
- Basturk O, Hong SM, Wood LD, Adsay NV, Albores-Saavedra J, Biankin AV, et al. A Revised Classification System and Recommendations From the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am J Surg Pathol 2015;39(12):1730–41 doi 10.1097/Pas.0000000000000533. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

