Structural and computational design of a SARS-CoV-2 spike antigen with improved expression and immunogenicity
- PMID: 37285422
- PMCID: PMC10246912
- DOI: 10.1126/sciadv.adg0330
Structural and computational design of a SARS-CoV-2 spike antigen with improved expression and immunogenicity
Abstract
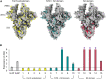
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern challenge the efficacy of approved vaccines, emphasizing the need for updated spike antigens. Here, we use an evolutionary-based design aimed at boosting protein expression levels of S-2P and improving immunogenic outcomes in mice. Thirty-six prototype antigens were generated in silico and 15 were produced for biochemical analysis. S2D14, which contains 20 computationally designed mutations within the S2 domain and a rationally engineered D614G mutation in the SD2 domain, has an ~11-fold increase in protein yield and retains RBD antigenicity. Cryo-electron microscopy structures reveal a mixture of populations in various RBD conformational states. Vaccination of mice with adjuvanted S2D14 elicited higher cross-neutralizing antibody titers than adjuvanted S-2P against the SARS-CoV-2 Wuhan strain and four variants of concern. S2D14 may be a useful scaffold or tool for the design of future coronavirus vaccines, and the approaches used for the design of S2D14 may be broadly applicable to streamline vaccine discovery.
Figures




References
-
- E. Volz, S. Mishra, M. Chand, J. C. Barrett, R. Johnson, L. Geidelberg, W. R. Hinsley, D. J. Laydon, G. Dabrera, Á. O’Toole, R. Amato, M. Ragonnet-Cronin, I. Harrison, B. Jackson, C. V. Ariani, O. Boyd, N. J. Loman, J. T. McCrone, S. Gonçalves, D. Jorgensen, R. Myers, V. Hill, D. K. Jackson, K. Gaythorpe, N. Groves, J. Sillitoe, D. P. Kwiatkowski; COVID-19 Genomics UK (COG-UK) consortium, S. Flaxman, O. Ratmann, S. Bhatt, S. Hopkins, A. Gandy, A. Rambaut, N. M. Ferguson, Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269 (2021). - PubMed
-
- H. Tegally, E. Wilkinson, M. Giovanetti, A. Iranzadeh, V. Fonseca, J. Giandhari, D. Doolabh, S. Pillay, E. J. San, N. Msomi, K. Mlisana, A. von Gottberg, S. Walaza, M. Allam, A. Ismail, T. Mohale, A. J. Glass, S. Engelbrecht, G. van Zyl, W. Preiser, F. Petruccione, A. Sigal, D. Hardie, G. Marais, N. Y. Hsiao, S. Korsman, M. A. Davies, L. Tyers, I. Mudau, D. York, C. Maslo, D. Goedhals, S. Abrahams, O. Laguda-Akingba, A. Alisoltani-Dehkordi, A. Godzik, C. K. Wibmer, B. T. Sewell, J. Lourenço, L. C. J. Alcantara, S. L. Kosakovsky Pond, S. Weaver, D. Martin, R. J. Lessells, J. N. Bhiman, C. Williamson, T. de Oliveira, Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592, 438–443 (2021). - PubMed
-
- S. Jangra, C. Ye, R. Rathnasinghe, D. Stadlbauer, F. Krammer, V. Simon, L. Martinez-Sobrido, A. García-Sastre, M. Schotsaert, H. Alshammary, A. A. Amoako, M. H. Awawda, K. F. Beach, M. C. Bermúdez-González, R. L. Chernet, L. Q. Eaker, E. D. Ferreri, D. L. Floda, C. R. Gleason, G. Kleiner, D. Jurczyszak, J. C. Matthews, W. A. Mendez, L. C. F. Mulder, K. T. Russo, A. B. T. Salimbangon, M. Saksena, A. S. Shin, L. A. Sominsky, K. Srivastava, SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2, e283–e284 (2021). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

