Human SAMD9 is a poxvirus-activatable anticodon nuclease inhibiting codon-specific protein synthesis
- PMID: 37285440
- PMCID: PMC10246899
- DOI: 10.1126/sciadv.adh8502
Human SAMD9 is a poxvirus-activatable anticodon nuclease inhibiting codon-specific protein synthesis
Abstract
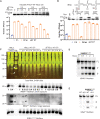
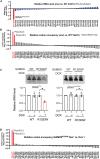
As a defense strategy against viruses or competitors, some microbes use anticodon nucleases (ACNases) to deplete essential tRNAs, effectively halting global protein synthesis. However, this mechanism has not been observed in multicellular eukaryotes. Here, we report that human SAMD9 is an ACNase that specifically cleaves phenylalanine tRNA (tRNAPhe), resulting in codon-specific ribosomal pausing and stress signaling. While SAMD9 ACNase activity is normally latent in cells, it can be activated by poxvirus infection or rendered constitutively active by SAMD9 mutations associated with various human disorders, revealing tRNAPhe depletion as an antiviral mechanism and a pathogenic condition in SAMD9 disorders. We identified the N-terminal effector domain of SAMD9 as the ACNase, with substrate specificity primarily determined by a eukaryotic tRNAPhe-specific 2'-O-methylation at the wobble position, making virtually all eukaryotic tRNAPhe susceptible to SAMD9 cleavage. Notably, the structure and substrate specificity of SAMD9 ACNase differ from known microbial ACNases, suggesting convergent evolution of a common immune defense strategy targeting tRNAs.
Figures




References
-
- H. Masaki, T. Ogawa, The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie 84, 433–438 (2002). - PubMed
-
- G. Kaufmann, Anticodon nucleases. Trends Biochem. Sci. 25, 70–74 (2000). - PubMed
-
- D. M. Thompson, R. Parker, Stressing out over tRNA cleavage. Cell 138, 215–219 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

