Mechanism and evolution of human ACE2 binding by SARS-CoV-2 spike
- PMID: 37285618
- PMCID: PMC10183628
- DOI: 10.1016/j.sbi.2023.102619
Mechanism and evolution of human ACE2 binding by SARS-CoV-2 spike
Abstract
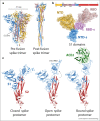
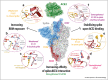
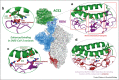
Spike glycoprotein of SARS-CoV-2 mediates viral entry into host cells by facilitating virus attachment and membrane fusion. ACE2 is the main receptor of SARS-CoV-2 and its interaction with spike has shaped the virus' emergence from an animal reservoir and subsequent evolution in the human host. Many structural studies on the spike:ACE2 interaction have provided insights into mechanisms driving viral evolution during the on-going pandemic. This review describes the molecular basis of spike binding to ACE2, outlines mechanisms that have optimised this interaction during viral evolution, and suggests directions for future research.
Copyright © 2023 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Figures

References
-
- Lau S.K.P., et al. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East Respiratory Syndrome Coronavirus. J Virol. 2013;87:8638–8650. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

