The plant cytosolic m6A RNA methylome stabilizes photosynthesis in the cold
- PMID: 37287225
- PMCID: PMC10721483
- DOI: 10.1016/j.xplc.2023.100634
The plant cytosolic m6A RNA methylome stabilizes photosynthesis in the cold
Abstract
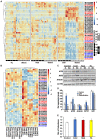
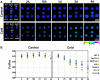
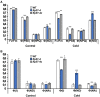
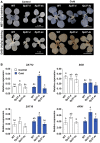
The sessile lifestyle of plants requires an immediate response to environmental stressors that affect photosynthesis, growth, and crop yield. Here, we showed that three abiotic perturbations-heat, cold, and high light-triggered considerable changes in the expression signatures of 42 epitranscriptomic factors (writers, erasers, and readers) with putative chloroplast-associated functions that formed clusters of commonly expressed genes in Arabidopsis. The expression changes under all conditions were reversible upon deacclimation, identifying epitranscriptomic players as modulators in acclimation processes. Chloroplast dysfunctions, particularly those induced by the oxidative stress-inducing norflurazon in a largely GENOME UNCOUPLED-independent manner, triggered retrograde signals to remodel chloroplast-associated epitranscriptomic expression patterns. N6-methyladenosine (m6A) is known as the most prevalent RNA modification and impacts numerous developmental and physiological functions in living organisms. During cold treatment, expression of components of the primary nuclear m6A methyltransferase complex was upregulated, accompanied by a significant increase in cellular m6A mRNA marks. In the cold, the presence of FIP37, a core component of the writer complex, played an important role in positive regulation of thylakoid structure, photosynthetic functions, and accumulation of photosystem I, the Cytb6f complex, cyclic electron transport proteins, and Curvature Thylakoid1 but not that of photosystem II components and the chloroplast ATP synthase. Downregulation of FIP37 affected abundance, polysomal loading, and translation of cytosolic transcripts related to photosynthesis in the cold, suggesting m6A-dependent translational regulation of chloroplast functions. In summary, we identified multifaceted roles of the cellular m6A RNA methylome in coping with cold; these were predominantly associated with chloroplasts and served to stabilize photosynthesis.
Keywords: Arabidopsis thaliana; RNA methylation; chloroplast; cold acclimation; m(6)A; photosynthesis; stress response.
Copyright © 2023 LMU Munich. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Armbruster U., Labs M., Pribil M., Viola S., Xu W., Scharfenberg M., Hertle A., Rojahn U., Jensen P., Rappaport F., et al. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell. 2013;25:2661–2678. doi: 10.1105/tpc.113.113118. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

