Identifying fibrogenic cells following salivary gland obstructive injury
- PMID: 37287453
- PMCID: PMC10242138
- DOI: 10.3389/fcell.2023.1190386
Identifying fibrogenic cells following salivary gland obstructive injury
Abstract
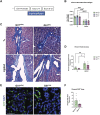
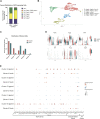
Fibrosis results from excess extracellular matrix accumulation, which alters normal tissue architecture and impedes function. In the salivary gland, fibrosis can be induced by irradiation treatment for cancer therapy, Sjögren's Disease, and other causes; however, it is unclear which stromal cells and signals participate in injury responses and disease progression. As hedgehog signaling has been implicated in fibrosis of the salivary gland and other organs, we examined contributions of the hedgehog effector, Gli1, to fibrotic responses in salivary glands. To experimentally induce a fibrotic response in female murine submandibular salivary glands, we performed ductal ligation surgery. We detected a progressive fibrotic response where both extracellular matrix accumulation and actively remodeled collagen significantly increased at 14 days post-ligation. Macrophages, which participate in extracellular matrix remodeling, and Gli1+ and PDGFRα+ stromal cells, which may deposit extracellular matrix, both increased with injury. Using single-cell RNA-sequencing, Gli1 + cells were not found in discrete clusters at embryonic day 16 but were found in clusters expressing the stromal genes Pdgfra and/or Pdgfrb. In adult mice, Gli1+ cells were similarly heterogeneous but more cells co-expressed PDGFRα and PDGFRβ. Using Gli1-CreERT2; ROSA26tdTomato lineage-tracing mice, we found that Gli1-derived cells expand with ductal ligation injury. Although some of the Gli1 lineage-traced tdTomato+ cells expressed vimentin and PDGFRβ following injury, there was no increase in the classic myofibroblast marker, smooth muscle alpha-actin. Additionally, there was little change in extracellular matrix area, remodeled collagen area, PDGFRα, PDGFRβ, endothelial cells, neurons, or macrophages in Gli1 null salivary glands following injury when compared with controls, suggesting that Gli1 signaling and Gli1+ cells have only a minor contribution to mechanical injury-induced fibrotic changes in the salivary gland. We used scRNA-seq to examine cell populations that expand with ligation and/or showed increased expression of matrisome genes. Some Pdgfra + /Pdgfrb + stromal cell subpopulations expanded in response to ligation, with two stromal cell subpopulations showing increased expression of Col1a1 and a greater diversity of matrisome genes, consistent with these cells being fibrogenic. However, only a few cells in these subpopulations expressed Gli1, consistent with a minor contribution of these cells to extracellular matrix production. Defining the signaling pathways driving fibrotic responses in stromal cell sub-types could reveal future therapeutic targets.
Keywords: Gli1; collagen; extracellular matrix (ECM); fibrosis; platelet derived growth factor receptor (PDGFR); salivary gland.
Copyright © 2023 Altrieth, O’Keefe, Gellatly, Tavarez, Feminella, Moskwa, Cordi, Turrieta, Nelson and Larsen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Update of
-
Identifying Fibrogenic Cells Following Salivary Gland Obstructive Injury.bioRxiv [Preprint]. 2023 Mar 10:2023.03.09.531751. doi: 10.1101/2023.03.09.531751. bioRxiv. 2023. Update in: Front Cell Dev Biol. 2023 May 23;11:1190386. doi: 10.3389/fcell.2023.1190386. PMID: 36945483 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

