Venetoclax durable response in adult relapsed/refractory Philadelphia-negative acute lymphoblastic leukemia with JAK/STAT pathway alterations
- PMID: 37287455
- PMCID: PMC10242111
- DOI: 10.3389/fcell.2023.1165308
Venetoclax durable response in adult relapsed/refractory Philadelphia-negative acute lymphoblastic leukemia with JAK/STAT pathway alterations
Abstract
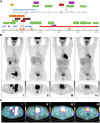
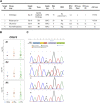
High-risk relapsed/refractory adult Philadelphia-negative (Ph-) B-cell acute lymphoblastic leukemia (B-ALL) is a great challenge due to limited possibilities to achieve and maintain a complete response. This also applies to cases with extramedullary (EM) involvement that have poor outcomes and no accepted standard therapeutic approaches. The incidence of EM localization in relapsed/refractory B-ALL is poorly investigated: data on patients treated with blinatumomab reported a 40% rate. Some responses were reported in EM patients with relapsed/refractory B-ALL treated with inotuzumab ozogamicin or CAR-T. However, molecular mechanisms of response or refractoriness are usually investigated neither at the medullary nor at EM sites. In the complex scenario of pluri-relapsed/refractory B-ALL patients, new target therapies are needed. Our analysis started with the case of an adult pluri-relapsed Ph- B-ALL patient, poorly sensitive to inotuzumab ozogamicin, donor lymphocyte infusions, and blinatumomab in EM disease, who achieved a durable/complete response after treatment with the BCL2-inhibitor venetoclax. The molecular characterization of medullary and EM samples revealed a tyrosine kinase domain JAK1 mutation in the bone marrow and EM samples at relapse. By comparing the expression level of BCL2- and JAK/STAT pathway-related genes between the patient samples, 136 adult JAK1 wt B-ALL, and 15 healthy controls, we identified differentially expressed genes, including LIFR, MTOR, SOCS1/2, and BCL2/BCL2L1, that are variably modulated at diverse time points and might explain the prolonged response to venetoclax (particularly in the EM site, which was only partially affected by previous therapies). Our results suggest that the deep molecular characterization of both medullary and EM samples is fundamental to identifying effective and personalized targeted therapies.
Keywords: JAK/STAT; Philadelphia-negative cells; acute lymphoblastic leukemia; extramedullary; venetoclax (BCL-2 inhibitor).
Copyright © 2023 Ferrari, Cangini, Ghelli Luserna di Rorà, Condorelli, Pugliese, Schininà, Cosentino, Fonzi, Domizio, Simonetti, Leotta, Milone and Martinelli.
Conflict of interest statement
GMa has competing interests with Novartis, BMS, Roche, Pfizer, ARIAD, and MSD. None of these agencies have had a role in the preparation of this manuscript. PCT application No. PCT/EP2021/065692 (10/06/2021): Method to identify linked genetic fusions. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Blink M., Buitenkamp T., Danen-van Oorschot A. A., de Haas V., Reinhardt D., Hasle H., et al. (2009). Mutation analysis of JAK-1, 2 and 3 in children with down Syndrome and acute leukemia. Blood 114, 5035. 10.1182/blood.v114.22.5035.5035 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

