Structural basis of the farnesoid X receptor/retinoid X receptor heterodimer on inverted repeat DNA
- PMID: 37287811
- PMCID: PMC10242635
- DOI: 10.1016/j.csbj.2023.05.026
Structural basis of the farnesoid X receptor/retinoid X receptor heterodimer on inverted repeat DNA
Abstract
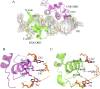
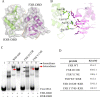
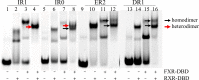
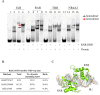
Farnesoid X receptor (FXR) is a ligand-activated transcription factor known as bile acid receptor (BAR). FXR plays critical roles in various biological processes, including metabolism, immune inflammation, liver regeneration and liver carcinogenesis. FXR forms a heterodimer with the retinoid X receptor (RXR) and binds to diverse FXR response elements (FXREs) to exert its various biological functions. However, the mechanism by which the FXR/RXR heterodimer binds the DNA elements remains unclear. In this study, we aimed to use structural, biochemical and bioinformatics analyses to study the mechanism of FXR binding to the typical FXRE, such as the IR1 site, and the heterodimer interactions in the FXR-DBD/RXR-DBD complex. Further biochemical assays showed that RAR, THR and NR4A2 do not form heterodimers with RXR when bound to the IR1 sites, which indicates that IR1 may be a unique binding site for the FXR/RXR heterodimer. Our studies may provide a further understanding of the dimerization specificity of nuclear receptors.
Keywords: Crystal structure; Farnesoid X receptor; Heterodimer; Inverted repeat DNA; Retinoid X receptor.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor.J Biol Chem. 2000 Apr 7;275(14):10638-47. doi: 10.1074/jbc.275.14.10638. J Biol Chem. 2000. PMID: 10744760
-
Retinoid X receptor (RXR) agonist-induced antagonism of farnesoid X receptor (FXR) activity due to absence of coactivator recruitment and decreased DNA binding.J Biol Chem. 2003 Mar 21;278(12):10028-32. doi: 10.1074/jbc.M208312200. Epub 2003 Jan 7. J Biol Chem. 2003. PMID: 12519787
-
Ligand binding and heterodimerization with retinoid X receptor α (RXRα) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding.J Biol Chem. 2018 Nov 23;293(47):18180-18191. doi: 10.1074/jbc.RA118.004652. Epub 2018 Oct 1. J Biol Chem. 2018. PMID: 30275017 Free PMC article.
-
Bile acid nuclear receptor FXR and digestive system diseases.Acta Pharm Sin B. 2015 Mar;5(2):135-44. doi: 10.1016/j.apsb.2015.01.004. Epub 2015 Feb 25. Acta Pharm Sin B. 2015. PMID: 26579439 Free PMC article. Review.
-
Bile salt excretory pump: biology and pathobiology.J Pediatr Gastroenterol Nutr. 2006 Jul;43 Suppl 1:S10-6. doi: 10.1097/01.mpg.0000226385.71859.5f. J Pediatr Gastroenterol Nutr. 2006. PMID: 16819395 Review.
Cited by
-
Nuclear receptor interdomain communication is mediated by the hinge with ligand specificity.bioRxiv [Preprint]. 2024 Apr 15:2024.02.10.579785. doi: 10.1101/2024.02.10.579785. bioRxiv. 2024. Update in: J Mol Biol. 2024 Nov 15;436(22):168805. doi: 10.1016/j.jmb.2024.168805. PMID: 38405809 Free PMC article. Updated. Preprint.
-
Recent Insights on the Role of Nuclear Receptors in Alzheimer's Disease: Mechanisms and Therapeutic Application.Int J Mol Sci. 2025 Jan 30;26(3):1207. doi: 10.3390/ijms26031207. Int J Mol Sci. 2025. PMID: 39940973 Free PMC article. Review.
-
Engineered bacteriophytochrome heterodimers for research and applications.J Biol Chem. 2025 Jul 4;301(8):110452. doi: 10.1016/j.jbc.2025.110452. Online ahead of print. J Biol Chem. 2025. PMID: 40617349 Free PMC article.
-
Retinoid X receptor heterodimers in hepatic function: structural insights and therapeutic potential.Front Pharmacol. 2024 Oct 16;15:1464655. doi: 10.3389/fphar.2024.1464655. eCollection 2024. Front Pharmacol. 2024. PMID: 39478961 Free PMC article. Review.
-
Structural basis for the asymmetric binding of coactivator SRC1 to FXR-RXRα and allosteric communication within the complex.Commun Biol. 2025 Mar 13;8(1):425. doi: 10.1038/s42003-025-07854-x. Commun Biol. 2025. PMID: 40082595 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
