Targeting NETs using dual-active DNase1 variants
- PMID: 37287977
- PMCID: PMC10242134
- DOI: 10.3389/fimmu.2023.1181761
Targeting NETs using dual-active DNase1 variants
Abstract
Background: Neutrophil Extracellular Traps (NETs) are key mediators of immunothrombotic mechanisms and defective clearance of NETs from the circulation underlies an array of thrombotic, inflammatory, infectious, and autoimmune diseases. Efficient NET degradation depends on the combined activity of two distinct DNases, DNase1 and DNase1-like 3 (DNase1L3) that preferentially digest double-stranded DNA (dsDNA) and chromatin, respectively.
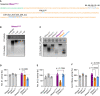
Methods: Here, we engineered a dual-active DNase with combined DNase1 and DNase1L3 activities and characterized the enzyme for its NET degrading potential in vitro. Furthermore, we produced a mouse model with transgenic expression of the dual-active DNase and analyzed body fluids of these animals for DNase1 and DNase 1L3 activities. We systematically substituted 20 amino acid stretches in DNase1 that were not conserved among DNase1 and DNase1L3 with homologous DNase1L3 sequences.
Results: We found that the ability of DNase1L3 to degrade chromatin is embedded into three discrete areas of the enzyme's core body, not the C-terminal domain as suggested by the state-of-the-art. Further, combined transfer of the aforementioned areas of DNase1L3 to DNase1 generated a dual-active DNase1 enzyme with additional chromatin degrading activity. The dual-active DNase1 mutant was superior to native DNase1 and DNase1L3 in degrading dsDNA and chromatin, respectively. Transgenic expression of the dual-active DNase1 mutant in hepatocytes of mice lacking endogenous DNases revealed that the engineered enzyme was stable in the circulation, released into serum and filtered to the bile but not into the urine.
Conclusion: Therefore, the dual-active DNase1 mutant is a promising tool for neutralization of DNA and NETs with potential therapeutic applications for interference with thromboinflammatory disease states.
Keywords: DNase1; DNase1-like 3; NET degradation; NETosis; neutrophil extracellular traps (NETs); protein engineering; recombinant proteins; thromboinflammation.
Copyright © 2023 Englert, Göbel, Khong, Omidi, Wolska, Konrath, Frye, Mailer, Beerens, Gerwers, Preston, Odeberg, Butler, Maas, Stavrou, Fuchs and Renné.
Conflict of interest statement
TF, JG, and HE hold a patent WO 2019/036719A2 “Engineered DNase Enzymes and Use in Therapy” licensed to Neutrolis Inc. TF is co-founder and CEO of Neutrolis Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

