Transcriptomic signatures reveal a shift towards an anti-inflammatory gene expression profile but also the induction of type I and type II interferon signaling networks through aryl hydrocarbon receptor activation in murine macrophages
- PMID: 37287978
- PMCID: PMC10242070
- DOI: 10.3389/fimmu.2023.1156493
Transcriptomic signatures reveal a shift towards an anti-inflammatory gene expression profile but also the induction of type I and type II interferon signaling networks through aryl hydrocarbon receptor activation in murine macrophages
Abstract
Introduction: The aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor that regulates a broad range of target genes involved in the xenobiotic response, cell cycle control and circadian rhythm. AhR is constitutively expressed in macrophages (Mϕ), acting as key regulator of cytokine production. While proinflammatory cytokines, i.e., IL-1β, IL-6, IL-12, are suppressed through AhR activation, anti-inflammatory IL-10 is induced. However, the underlying mechanisms of those effects and the importance of the specific ligand structure are not yet completely understood.
Methods: Therefore, we have compared the global gene expression pattern in activated murine bone marrow-derived macrophages (BMMs) subsequently to exposure with either benzo[a]pyrene (BaP) or indole-3-carbinol (I3C), representing high-affinity vs. low-affinity AhR ligands, respectively, by means of mRNA sequencing. AhR dependency of observed effects was proved using BMMs from AhR-knockout (Ahr-/-) mice.
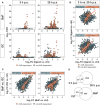
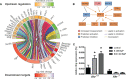
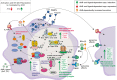
Results and discussion: In total, more than 1,000 differentially expressed genes (DEGs) could be mapped, covering a plethora of AhR-modulated effects on basal cellular processes, i.e., transcription and translation, but also immune functions, i.e., antigen presentation, cytokine production, and phagocytosis. Among DEGs were genes that are already known to be regulated by AhR, i.e., Irf1, Ido2, and Cd84. However, we identified DEGs not yet described to be AhR-regulated in Mϕ so far, i.e., Slpi, Il12rb1, and Il21r. All six genes likely contribute to shifting the Mϕ phenotype from proinflammatory to anti-inflammatory. The majority of DEGs induced through BaP were not affected through I3C exposure, probably due to higher AhR affinity of BaP in comparison to I3C. Mapping of known aryl hydrocarbon response element (AHRE) sequence motifs in identified DEGs revealed more than 200 genes not possessing any AHRE, and therefore being not eligible for canonical regulation. Bioinformatic approaches modeled a central role of type I and type II interferons in the regulation of those genes. Additionally, RT-qPCR and ELISA confirmed a AhR-dependent expressional induction and AhR-dependent secretion of IFN-γ in response to BaP exposure, suggesting an auto- or paracrine activation pathway of Mϕ.
Keywords: aryl hydrocarbon receptor; benzo[a]pyrene; immunomodulation; indol-3-carbinol; innate immunity; macrophage activation; transcriptomics; type I/II interferons.
Copyright © 2023 Schmidt, Haupt, Riemschneider, Kämpf, Löffler, Blumert, Reiche, Koehl, Kalkhof and Lehmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

