Structurally reconfigurable designer RNA structures for nanomachines
- PMID: 37288082
- PMCID: PMC10240537
- DOI: 10.52601/bpr.2021.200053
Structurally reconfigurable designer RNA structures for nanomachines
Abstract
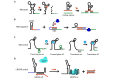
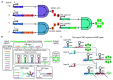
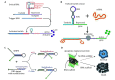
Structurally reconfigurable RNA structures enable dynamic transitions of the functional states in response to diverse molecular stimuli, which are fundamental in genetic and epigenetic regulations. Inspired by nature, rationally designed RNA structures with responsively reconfigurable motifs have been developed to serve as switchable components for building engineered nanomachines, which hold promise in synthetic biological applications. In this review, we summarize recent progress in the design, synthesis, and integration of engineered reconfigurable RNA structures for nanomachines. We highlight recent examples of their targeted applications such as biocomputing and smart theranostics. We also discuss their advantages, challenges as well as possible solutions. We further provide an outlook of their potential in future synthetic biology.
Keywords: Biocomputing; RNA nanomachine; Reconfiguration; Structure; Theranostics.
© The Author(s) 2021.
Conflict of interest statement
Kai Jiao, Yaya Hao, Fei Wang, Lihua Wang, Chunhai Fan and Jiang Li declare that they have no conflict of interest.
Figures



Similar articles
-
Smart Actuators and Adhesives for Reconfigurable Matter.Acc Chem Res. 2017 Apr 18;50(4):691-702. doi: 10.1021/acs.accounts.6b00612. Epub 2017 Mar 6. Acc Chem Res. 2017. PMID: 28263544
-
Materials and Schemes of Multimodal Reconfigurable Micro/Nanomachines and Robots: Review and Perspective.Adv Mater. 2021 Oct;33(39):e2101965. doi: 10.1002/adma.202101965. Epub 2021 Aug 19. Adv Mater. 2021. PMID: 34410023 Review.
-
Dynamic Reconfigurable DNA Nanostructures, Networks and Materials.Angew Chem Int Ed Engl. 2023 Apr 24;62(18):e202215332. doi: 10.1002/anie.202215332. Epub 2023 Feb 14. Angew Chem Int Ed Engl. 2023. PMID: 36651472 Review.
-
Protein cages, rings and tubes: useful components of future nanodevices?Nanotechnol Sci Appl. 2008 Nov 17;1:67-78. doi: 10.2147/nsa.s4092. Nanotechnol Sci Appl. 2008. PMID: 24198461 Free PMC article. Review.
-
Switchable reconfiguration of nucleic acid nanostructures by stimuli-responsive DNA machines.Acc Chem Res. 2014 Jun 17;47(6):1673-80. doi: 10.1021/ar400316h. Epub 2014 Mar 21. Acc Chem Res. 2014. PMID: 24654959
Cited by
-
Diversity of Self-Assembled RNA Complexes: From Nanoarchitecture to Nanomachines.Molecules. 2023 Dec 19;29(1):10. doi: 10.3390/molecules29010010. Molecules. 2023. PMID: 38202593 Free PMC article.
-
Optimized lipid nanoparticles (LNPs) for organ-selective nucleic acids delivery in vivo.iScience. 2024 Apr 23;27(6):109804. doi: 10.1016/j.isci.2024.109804. eCollection 2024 Jun 21. iScience. 2024. PMID: 38770138 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
