Catalytic Cycle of the Bifunctional Enzyme Phosphoribosyl-ATP Pyrophosphohydrolase/Phosphoribosyl-AMP Cyclohydrolase
- PMID: 37288093
- PMCID: PMC10242683
- DOI: 10.1021/acscatal.3c01111
Catalytic Cycle of the Bifunctional Enzyme Phosphoribosyl-ATP Pyrophosphohydrolase/Phosphoribosyl-AMP Cyclohydrolase
Abstract
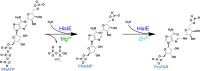
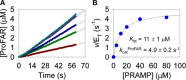
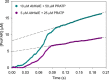
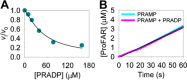
The bifunctional enzyme phosphoribosyl-ATP pyrophosphohydrolase/phosphoribosyl-AMP cyclohydrolase (HisIE) catalyzes the second and third steps of histidine biosynthesis: pyrophosphohydrolysis of N1-(5-phospho-β-D-ribosyl)-ATP (PRATP) to N1-(5-phospho-β-D-ribosyl)-AMP (PRAMP) and pyrophosphate in the C-terminal HisE-like domain, and cyclohydrolysis of PRAMP to N-(5'-phospho-D-ribosylformimino)-5-amino-1-(5″-phospho-D-ribosyl)-4-imidazolecarboxamide (ProFAR) in the N-terminal HisI-like domain. Here we use UV-VIS spectroscopy and LC-MS to show Acinetobacter baumannii putative HisIE produces ProFAR from PRATP. Employing an assay to detect pyrophosphate and another to detect ProFAR, we established the pyrophosphohydrolase reaction rate is higher than the overall reaction rate. We produced a truncated version of the enzyme-containing only the C-terminal (HisE) domain. This truncated HisIE was catalytically active, which allowed the synthesis of PRAMP, the substrate for the cyclohydrolysis reaction. PRAMP was kinetically competent for HisIE-catalyzed ProFAR production, demonstrating PRAMP can bind the HisI-like domain from bulk water, and suggesting that the cyclohydrolase reaction is rate-limiting for the overall bifunctional enzyme. The overall kcat increased with increasing pH, while the solvent deuterium kinetic isotope effect decreased at more basic pH but was still large at pH 7.5. The lack of solvent viscosity effects on kcat and kcat/KM ruled out diffusional steps limiting the rates of substrate binding and product release. Rapid kinetics with excess PRATP demonstrated a lag time followed by a burst in ProFAR formation. These observations are consistent with a rate-limiting unimolecular step involving a proton transfer following adenine ring opening. We synthesized N1-(5-phospho-β-D-ribosyl)-ADP (PRADP), which could not be processed by HisIE. PRADP inhibited HisIE-catalyzed ProFAR formation from PRATP but not from PRAMP, suggesting that it binds to the phosphohydrolase active site while still permitting unobstructed access of PRAMP to the cyclohydrolase active site. The kinetics data are incompatible with a build-up of PRAMP in bulk solvent, indicating HisIE catalysis involves preferential channeling of PRAMP, albeit not via a protein tunnel.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



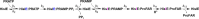
Similar articles
-
Crystal Structure, Steady-State, and Pre-Steady-State Kinetics of Acinetobacter baumannii ATP Phosphoribosyltransferase.Biochemistry. 2024 Jan 16;63(2):230-240. doi: 10.1021/acs.biochem.3c00551. Epub 2023 Dec 27. Biochemistry. 2024. PMID: 38150593 Free PMC article.
-
Isolation and characterization of a histidine biosynthetic gene in Arabidopsis encoding a polypeptide with two separate domains for phosphoribosyl-ATP pyrophosphohydrolase and phosphoribosyl-AMP cyclohydrolase.Plant Physiol. 1998 Sep;118(1):275-83. doi: 10.1104/pp.118.1.275. Plant Physiol. 1998. PMID: 9733547 Free PMC article.
-
Allosteric Activation Shifts the Rate-Limiting Step in a Short-Form ATP Phosphoribosyltransferase.Biochemistry. 2018 Jul 24;57(29):4357-4367. doi: 10.1021/acs.biochem.8b00559. Epub 2018 Jul 10. Biochemistry. 2018. PMID: 29940105 Free PMC article.
-
Biosynthesis of D-arabinose in mycobacteria - a novel bacterial pathway with implications for antimycobacterial therapy.FEBS J. 2008 Jun;275(11):2691-711. doi: 10.1111/j.1742-4658.2008.06395.x. Epub 2008 Apr 15. FEBS J. 2008. PMID: 18422659 Review.
-
The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia.Biochemistry. 1999 Jun 22;38(25):7891-9. doi: 10.1021/bi990871p. Biochemistry. 1999. PMID: 10387030 Review.
Cited by
-
Kinetic Dissection of the Reaction of Human GDP-l-Fucose Synthase.ACS Catal. 2025 Jul 29;15(16):13872-13885. doi: 10.1021/acscatal.5c02722. eCollection 2025 Aug 15. ACS Catal. 2025. PMID: 40837376 Free PMC article.
-
From Soil to Surface: Exploring the Impact of Green Infrastructure on Microbial Communities in the Built Environment.J Genomics. 2025 Jan 18;13:10-23. doi: 10.7150/jgen.106245. eCollection 2025. J Genomics. 2025. PMID: 39925382 Free PMC article.
-
Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.ACS Catal. 2024 Feb 13;14(5):3090-3102. doi: 10.1021/acscatal.3c06260. eCollection 2024 Mar 1. ACS Catal. 2024. PMID: 38449528 Free PMC article.
-
Crystal Structure, Steady-State, and Pre-Steady-State Kinetics of Acinetobacter baumannii ATP Phosphoribosyltransferase.Biochemistry. 2024 Jan 16;63(2):230-240. doi: 10.1021/acs.biochem.3c00551. Epub 2023 Dec 27. Biochemistry. 2024. PMID: 38150593 Free PMC article.
-
Allosteric activation unveils protein-mass modulation of ATP phosphoribosyltransferase product release.Commun Chem. 2024 Apr 6;7(1):77. doi: 10.1038/s42004-024-01165-8. Commun Chem. 2024. PMID: 38582930 Free PMC article.
References
-
- Martin R. G.; Berberich M. A.; Ames B. N.; Davis W. W.; Goldberger R. F.; Yourno J. D. Enzymes And Intermediates Of Histidine Biosynthesis In Salmonella Typhimurium. Methods Enzymol. 1971, 17, 3–44. 10.1016/0076-6879(71)17003-6. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
