Nuclear segmentation facilitates neutrophil migration
- PMID: 37288767
- PMCID: PMC10309577
- DOI: 10.1242/jcs.260768
Nuclear segmentation facilitates neutrophil migration
Abstract
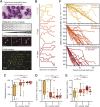
Neutrophils are among the fastest-moving immune cells. Their speed is critical to their function as 'first responder' cells at sites of damage or infection, and it has been postulated that the unique segmented nucleus of neutrophils functions to assist their rapid migration. Here, we tested this hypothesis by imaging primary human neutrophils traversing narrow channels in custom-designed microfluidic devices. Individuals were given an intravenous low dose of endotoxin to elicit recruitment of neutrophils into the blood with a high diversity of nuclear phenotypes, ranging from hypo- to hyper-segmented. Both by cell sorting of neutrophils from the blood using markers that correlate with lobularity and by directly quantifying the migration of neutrophils with distinct lobe numbers, we found that neutrophils with one or two nuclear lobes were significantly slower to traverse narrower channels, compared to neutrophils with more than two nuclear lobes. Thus, our data show that nuclear segmentation in primary human neutrophils provides a speed advantage during migration through confined spaces.
Keywords: Endotoxemia; Microfluidics; Migration; Neutrophil; Nuclear segmentation; Nucleus.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

