Characterization of an Emergent Chicken H3N8 Influenza Virus in Southern China: a Potential Threat to Public Health
- PMID: 37289052
- PMCID: PMC10308888
- DOI: 10.1128/jvi.00434-23
Characterization of an Emergent Chicken H3N8 Influenza Virus in Southern China: a Potential Threat to Public Health
Abstract
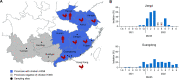
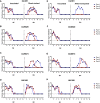
Although influenza A viruses of several subtypes have occasionally infected humans, to date only those of the H1, H2, and H3 subtypes have led to pandemics and become established in humans. The detection of two human infections by avian H3N8 viruses in April and May of 2022 raised pandemic concerns. Recent studies have shown the H3N8 viruses were introduced into humans from poultry, although their genesis, prevalence, and transmissibility in mammals have not been fully elucidated. Findings generated from our systematic influenza surveillance showed that this H3N8 influenza virus was first detected in chickens in July 2021 and then disseminated and became established in chickens over wider regions of China. Phylogenetic analyses revealed that the H3 HA and N8 NA were derived from avian viruses prevalent in domestic ducks in the Guangxi-Guangdong region, while all internal genes were from enzootic poultry H9N2 viruses. The novel H3N8 viruses form independent lineages in the glycoprotein gene trees, but their internal genes are mixed with those of H9N2 viruses, indicating continuous gene exchange among these viruses. Experimental infection of ferrets with three chicken H3N8 viruses showed transmission through direct contact and inefficient transmission by airborne exposure. Examination of contemporary human sera detected only very limited antibody cross-reaction to these viruses. The continuing evolution of these viruses in poultry could pose an ongoing pandemic threat. IMPORTANCE A novel H3N8 virus with demonstrated zoonotic potential has emerged and disseminated in chickens in China. It was generated by reassortment between avian H3 and N8 virus(es) and long-term enzootic H9N2 viruses present in southern China. This H3N8 virus has maintained independent H3 and N8 gene lineages but continues to exchange internal genes with other H9N2 viruses to form novel variants. Our experimental studies showed that these H3N8 viruses were transmissible in ferrets, and serological data suggest that the human population lacks effective immunological protection against it. With its wide geographical distribution and continuing evolution in chickens, other spillovers to humans can be expected and might lead to more efficient transmission in humans.
Keywords: H3N8; chicken; ferret; infectivity; influenza; pathogenesis; transmissibility; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi:10.1126/science.1176225. - DOI - PMC - PubMed
-
- Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X, Li L, Hong W, Chai Y, Zhou L, Liang H, Ou Z, Liu Y, Farooqui A, Kelvin DJ, Poon LL, Smith DK, Pybus OG, Leung GM, Shu Y, Webster RG, Webby RJ, Peiris JS, Rambaut A, Zhu H, Guan Y. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502:241–244. doi:10.1038/nature12515. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

