Microbiomes of Blood-Feeding Triatomines in the Context of Their Predatory Relatives and the Environment
- PMID: 37289079
- PMCID: PMC10433993
- DOI: 10.1128/spectrum.01681-23
Microbiomes of Blood-Feeding Triatomines in the Context of Their Predatory Relatives and the Environment
Abstract
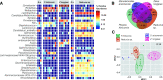
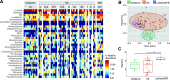
The importance of gut microbiomes has become generally recognized in vector biology. This study addresses microbiome signatures in North American Triatoma species of public health significance (vectors of Trypanosoma cruzi) linked to their blood-feeding strategy and the natural habitat. To place the Triatoma-associated microbiomes within a complex evolutionary and ecological context, we sampled sympatric Triatoma populations, related predatory reduviids, unrelated ticks, and environmental material from vertebrate nests where these arthropods reside. Along with five Triatoma species, we have characterized microbiomes of five reduviids (Stenolemoides arizonensis, Ploiaria hirticornis, Zelus longipes, and two Reduvius species), a single soft tick species, Ornithodoros turicata, and environmental microbiomes from selected sites in Arizona, Texas, Florida, and Georgia. The microbiomes of predatory reduviids lack a shared core microbiota. As in triatomines, microbiome dissimilarities among species correlate with dominance of a single bacterial taxon. These include Rickettsia, Lactobacillus, "Candidatus Midichloria," and Zymobacter, which are often accompanied by known symbiotic genera, i.e., Wolbachia, "Candidatus Lariskella," Asaia, Gilliamella, and Burkholderia. We have further identified a compositional convergence of the analyzed microbiomes in regard to the host phylogenetic distance in both blood-feeding and predatory reduviids. While the microbiomes of the two reduviid species from the Emesinae family reflect their close relationship, the microbiomes of all Triatoma species repeatedly form a distinct monophyletic cluster highlighting their phylosymbiosis. Furthermore, based on environmental microbiome profiles and blood meal analysis, we propose three epidemiologically relevant and mutually interrelated bacterial sources for Triatoma microbiomes, i.e., host abiotic environment, host skin microbiome, and pathogens circulating in host blood. IMPORTANCE This study places microbiomes of blood-feeding North American Triatoma vectors (Reduviidae) into a broader evolutionary and ecological context provided by related predatory assassin bugs (Reduviidae), another unrelated vector species (soft tick Ornithodoros turicata), and the environment these arthropods coinhabit. For both vectors, microbiome analyses suggest three interrelated sources of bacteria, i.e., the microbiome of vertebrate nests as their natural habitat, the vertebrate skin microbiome, and the pathobiome circulating in vertebrate blood. Despite an apparent influx of environment-associated bacteria into the arthropod microbiomes, Triatoma microbiomes retain their specificity, forming a distinct cluster that significantly differs from both predatory relatives and ecologically comparable ticks. Similarly, within the related predatory Reduviidae, we found the host phylogenetic distance to underlie microbiome similarities.
Keywords: Ornithodoros; Reduviidae; Triatoma; environment; microbiome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Perotti MA, Kirkness EF, Reed DL, Braig HR. 2008. Endosymbionts of lice, p 205–220. In Bourtzis K, Miller TA (ed), Insect symbiosis, 1st ed, vol 3. CRC Press, Boca Raton, FL.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

