Multiscale model of the different modes of cancer cell invasion
- PMID: 37289551
- PMCID: PMC10293590
- DOI: 10.1093/bioinformatics/btad374
Multiscale model of the different modes of cancer cell invasion
Abstract
Motivation: Mathematical models of biological processes altered in cancer are built using the knowledge of complex networks of signaling pathways, detailing the molecular regulations inside different cell types, such as tumor cells, immune and other stromal cells. If these models mainly focus on intracellular information, they often omit a description of the spatial organization among cells and their interactions, and with the tumoral microenvironment.
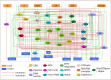
Results: We present here a model of tumor cell invasion simulated with PhysiBoSS, a multiscale framework, which combines agent-based modeling and continuous time Markov processes applied on Boolean network models. With this model, we aim to study the different modes of cell migration and to predict means to block it by considering not only spatial information obtained from the agent-based simulation but also intracellular regulation obtained from the Boolean model.
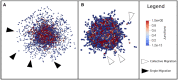
Our multiscale model integrates the impact of gene mutations with the perturbation of the environmental conditions and allows the visualization of the results with 2D and 3D representations. The model successfully reproduces single and collective migration processes and is validated on published experiments on cell invasion. In silico experiments are suggested to search for possible targets that can block the more invasive tumoral phenotypes.
Availability and implementation: https://github.com/sysbio-curie/Invasion_model_PhysiBoSS.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


References
-
- Anastas JN, Moon RT.. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013;13:11–26. - PubMed
-
- Anderson ARA. A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion. Math Med Biol 2005;22:163–86. - PubMed
-
- Anderson ARA. A hybrid multiscale model of solid tumour growth and invasion: evolution and the microenvironment. In: Anderson ARA, Chaplain MAJ, Rejniak KA (eds), Single-Cell-Based Models in Biology and Medicine, Mathematics and Biosciences in Interaction. Basel: Birkhäuser, 2007, 3–28.

