Combining DNA scaffolds and acoustic force spectroscopy to characterize individual protein bonds
- PMID: 37290437
- PMCID: PMC10323022
- DOI: 10.1016/j.bpj.2023.05.004
Combining DNA scaffolds and acoustic force spectroscopy to characterize individual protein bonds
Abstract
Single-molecule data are of great significance in biology, chemistry, and medicine. However, new experimental tools to characterize, in a multiplexed manner, protein bond rupture under force are still needed. Acoustic force spectroscopy is an emerging manipulation technique which generates acoustic waves to apply force in parallel on multiple microbeads tethered to a surface. We here exploit this configuration in combination with the recently developed modular junctured-DNA scaffold that has been designed to study protein-protein interactions at the single-molecule level. By applying repetitive constant force steps on the FKBP12-rapamycin-FRB complex, we measure its unbinding kinetics under force at the single-bond level. Special efforts are made in analyzing the data to identify potential pitfalls. We propose a calibration method allowing in situ force determination during the course of the unbinding measurement. We compare our results with well-established techniques, such as magnetic tweezers, to ensure their accuracy. We also apply our strategy to study the force-dependent rupture of a single-domain antibody with its antigen. Overall, we get a good agreement with the published parameters that have been obtained at zero force and population level. Thus, our technique offers single-molecule precision for multiplexed measurements of interactions of biotechnological and medical interest.
Copyright © 2023 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests PSL valorisation has submitted a patent related to the J-DNA (PCT FR2018/053533) with D.K., T.S., and C.G. among the inventors.
Figures

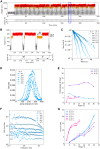
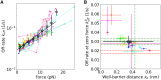
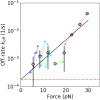
Similar articles
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
Cited by
-
Modulation of SARS-CoV-2 spike binding to ACE2 through conformational selection.Nat Nanotechnol. 2025 Jul;20(7):926-934. doi: 10.1038/s41565-025-01908-1. Epub 2025 Jun 10. Nat Nanotechnol. 2025. PMID: 40494932
-
Unravelling the Biophysical Properties of Chromatin Proteins and DNA Using Acoustic Force Spectroscopy.Methods Mol Biol. 2024;2819:519-534. doi: 10.1007/978-1-0716-3930-6_24. Methods Mol Biol. 2024. PMID: 39028522
References
-
- Limozin L., Puech P.-H. Membrane organization and physical regulation of lymphocyte antigen receptors: a biophysicist’s perspective. J. Membr. Biol. 2019;252:397–412. - PubMed

