South Asian medical cohorts reveal strong founder effects and high rates of homozygosity
- PMID: 37291107
- PMCID: PMC10250394
- DOI: 10.1038/s41467-023-38766-1
South Asian medical cohorts reveal strong founder effects and high rates of homozygosity
Abstract
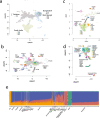
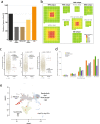
The benefits of large-scale genetic studies for healthcare of the populations studied are well documented, but these genetic studies have traditionally ignored people from some parts of the world, such as South Asia. Here we describe whole genome sequence (WGS) data from 4806 individuals recruited from the healthcare delivery systems of Pakistan, India and Bangladesh, combined with WGS from 927 individuals from isolated South Asian populations. We characterize population structure in South Asia and describe a genotyping array (SARGAM) and imputation reference panel that are optimized for South Asian genomes. We find evidence for high rates of reproductive isolation, endogamy and consanguinity that vary across the subcontinent and that lead to levels of rare homozygotes that reach 100 times that seen in outbred populations. Founder effects increase the power to associate functional variants with disease processes and make South Asia a uniquely powerful place for population-scale genetic studies.
© 2023. The Author(s).
Conflict of interest statement
J.D.W. has worked as a consultant for Genentech, MedGenome, and Maze Therapeutics, and has received research funding from Genentech. J.F.S. is a former employee and shareholder of 23andMe. A.V.K. is an employee of Verve Therapeutics; has served as a scientific advisor for Sanofi, Amgen, Maze Therapeutics, Novartis, Silence Therapeutics, Veritas International, Color Health, and Third Rock Ventures; holds equity in Verve Therapeutics, Marea Therapeutics, Color Health and Foresite Labs; and received a sponsored research agreement from IBM Research. The remaining authors declare no competing interests.
Figures