Combining SNAPs with antibiotics shows enhanced synergistic efficacy against S. aureus and P. aeruginosa biofilms
- PMID: 37291132
- PMCID: PMC10250483
- DOI: 10.1038/s41522-023-00401-8
Combining SNAPs with antibiotics shows enhanced synergistic efficacy against S. aureus and P. aeruginosa biofilms
Erratum in
-
Author Correction: Combining SNAPs with antibiotics shows enhanced synergistic efficacy against S. aureus and P. aeruginosa biofilms.NPJ Biofilms Microbiomes. 2023 Jun 16;9(1):38. doi: 10.1038/s41522-023-00407-2. NPJ Biofilms Microbiomes. 2023. PMID: 37328534 Free PMC article. No abstract available.
Abstract
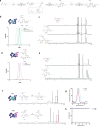
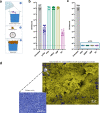
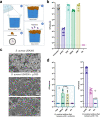
Biofilm infections are associated with a high mortality risk for patients. Antibiotics perform poorly against biofilm communities, so high doses and prolonged treatments are often used in clinical settings. We investigated the pairwise interactions of two synthetic nano-engineered antimicrobial polymers (SNAPs). The g-D50 copolymer was synergistic with penicillin and silver sulfadiazine against planktonic Staphylococcus aureus USA300 in synthetic wound fluid. Furthermore, the combination of g-D50 and silver sulfadiazine showed a potent synergistic antibiofilm activity against S. aureus USA300 using in vitro and ex vivo wound biofilm models. The a-T50 copolymer was synergistic with colistin against planktonic Pseudomonas aeruginosa in synthetic cystic fibrosis medium, and this pair showed a potent synergistic antibiofilm activity against P. aeruginosa in an ex vivo cystic fibrosis lung model. SNAPs thus have the potential for increased antibiofilm performance in combination with certain antibiotics to shorten prolonged treatments and reduce dosages against biofilm infection.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

