Astrocyte-oligodendrocyte interaction regulates central nervous system regeneration
- PMID: 37291151
- PMCID: PMC10250470
- DOI: 10.1038/s41467-023-39046-8
Astrocyte-oligodendrocyte interaction regulates central nervous system regeneration
Abstract
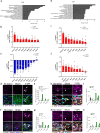
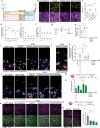
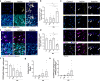
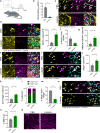
Failed regeneration of myelin around neuronal axons following central nervous system damage contributes to nerve dysfunction and clinical decline in various neurological conditions, for which there is an unmet therapeutic demand. Here, we show that interaction between glial cells - astrocytes and mature myelin-forming oligodendrocytes - is a determinant of remyelination. Using in vivo/ ex vivo/ in vitro rodent models, unbiased RNA sequencing, functional manipulation, and human brain lesion analyses, we discover that astrocytes support the survival of regenerating oligodendrocytes, via downregulation of the Nrf2 pathway associated with increased astrocytic cholesterol biosynthesis pathway activation. Remyelination fails following sustained astrocytic Nrf2 activation in focally-lesioned male mice yet is restored by either cholesterol biosynthesis/efflux stimulation, or Nrf2 inhibition using the existing therapeutic Luteolin. We identify that astrocyte-oligodendrocyte interaction regulates remyelination, and reveal a drug strategy for central nervous system regeneration centred on targeting this interaction.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

