RNA activation in ticks
- PMID: 37291173
- PMCID: PMC10250327
- DOI: 10.1038/s41598-023-36523-4
RNA activation in ticks
Abstract
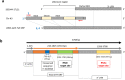
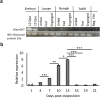
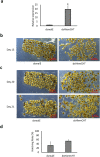
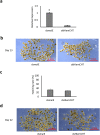
RNA activation (RNAa) is a burgeoning area of research in which double-stranded RNAs (dsRNAs) or small activating RNAs mediate the upregulation of specific genes by targeting the promoter sequence and/or AU-rich elements in the 3'- untranslated region (3'-UTR) of mRNA molecules. So far, studies on the phenomenon have been limited to mammals, plants, bacteria, Caenorhabditis elegans, and recently, Aedes aegypti. However, it is yet to be applied in other arthropods, including ticks, despite the ubiquitous presence of argonaute 2 protein, which is an indispensable requirement for the formation of RNA-induced transcriptional activation complex to enable a dsRNA-mediated gene activation. In this study, we demonstrated for the first time the possible presence of RNAa phenomenon in the tick vector, Haemaphysalis longicornis (Asian longhorned tick). We targeted the 3'-UTR of a novel endochitinase-like gene (HlemCHT) identified previously in H. longicornis eggs for dsRNA-mediated gene activation. Our results showed an increased gene expression in eggs of H. longicornis endochitinase-dsRNA-injected (dsHlemCHT) ticks on day-13 post-oviposition. Furthermore, we observed that eggs of dsHlemCHT ticks exhibited relatively early egg development and hatching, suggesting a dsRNA-mediated activation of the HlemCHT gene in the eggs. This is the first attempt to provide evidence of RNAa in ticks. Although further studies are required to elucidate the detailed mechanism by which RNAa occurs in ticks, the outcome of this study provides new opportunities for the use of RNAa as a gene overexpression tool in future studies on tick biology, to reduce the global burden of ticks and tick-borne diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Zhao, X. Y., Voutila, J., Habib, N. A. & Reebye, V. RNA Activation. Innov. Med. 241–249. 10.1007/978-4-431-55651-0_20 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

