Integrated multi-omics for rapid rare disease diagnosis on a national scale
- PMID: 37291213
- PMCID: PMC10353936
- DOI: 10.1038/s41591-023-02401-9
Integrated multi-omics for rapid rare disease diagnosis on a national scale
Abstract
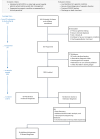
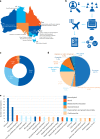
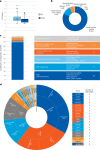
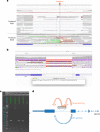
Critically ill infants and children with rare diseases need equitable access to rapid and accurate diagnosis to direct clinical management. Over 2 years, the Acute Care Genomics program provided whole-genome sequencing to 290 families whose critically ill infants and children were admitted to hospitals throughout Australia with suspected genetic conditions. The average time to result was 2.9 d and diagnostic yield was 47%. We performed additional bioinformatic analyses and transcriptome sequencing in all patients who remained undiagnosed. Long-read sequencing and functional assays, ranging from clinically accredited enzyme analysis to bespoke quantitative proteomics, were deployed in selected cases. This resulted in an additional 19 diagnoses and an overall diagnostic yield of 54%. Diagnostic variants ranged from structural chromosomal abnormalities through to an intronic retrotransposon, disrupting splicing. Critical care management changed in 120 diagnosed patients (77%). This included major impacts, such as informing precision treatments, surgical and transplant decisions and palliation, in 94 patients (60%). Our results provide preliminary evidence of the clinical utility of integrating multi-omic approaches into mainstream diagnostic practice to fully realize the potential of rare disease genomic testing in a timely manner.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Comment in
-
A rapid whole-genome sequencing service for infants with rare diseases in the United Arab Emirates.Nat Med. 2023 Dec;29(12):2979-2980. doi: 10.1038/s41591-023-02596-x. Nat Med. 2023. PMID: 37872224 No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

