B cell profiles, antibody repertoire and reactivity reveal dysregulated responses with autoimmune features in melanoma
- PMID: 37291228
- PMCID: PMC10249578
- DOI: 10.1038/s41467-023-39042-y
B cell profiles, antibody repertoire and reactivity reveal dysregulated responses with autoimmune features in melanoma
Abstract
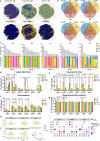
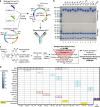
B cells are known to contribute to the anti-tumor immune response, especially in immunogenic tumors such as melanoma, yet humoral immunity has not been characterized in these cancers to detail. Here we show comprehensive phenotyping in samples of circulating and tumor-resident B cells as well as serum antibodies in melanoma patients. Memory B cells are enriched in tumors compared to blood in paired samples and feature distinct antibody repertoires, linked to specific isotypes. Tumor-associated B cells undergo clonal expansion, class switch recombination, somatic hypermutation and receptor revision. Compared with blood, tumor-associated B cells produce antibodies with proportionally higher levels of unproductive sequences and distinct complementarity determining region 3 properties. The observed features are signs of affinity maturation and polyreactivity and suggest an active and aberrant autoimmune-like reaction in the tumor microenvironment. Consistent with this, tumor-derived antibodies are polyreactive and characterized by autoantigen recognition. Serum antibodies show reactivity to antigens attributed to autoimmune diseases and cancer, and their levels are higher in patients with active disease compared to post-resection state. Our findings thus reveal B cell lineage dysregulation with distinct antibody repertoire and specificity, alongside clonally-expanded tumor-infiltrating B cells with autoimmune-like features, shaping the humoral immune response in melanoma.
© 2023. The Author(s).
Conflict of interest statement
S.N.K. and J.S. are founders and shareholders of Epsilogen Ltd. H.J.B. is employed through a fund provided by Epsilogen Ltd. S.N.K., J.S., and H.J.B. are inventors of patents on antibody technologies. All other authors have declared no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

