The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms
- PMID: 37291396
- PMCID: PMC10546791
- DOI: 10.1038/s41477-023-01435-8
The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms
Abstract
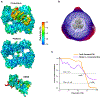
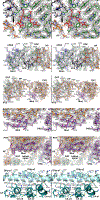
Plants employ a divergent cohort of phytochrome (Phy) photoreceptors to govern many aspects of morphogenesis through reversible photointerconversion between inactive Pr and active Pfr conformers. The two most influential are PhyA whose retention of Pfr enables sensation of dim light, while the relative instability of Pfr for PhyB makes it better suited for detecting full sun and temperature. To better understand these contrasts, we solved, by cryo-electron microscopy, the three-dimensional structure of full-length PhyA as Pr. Like PhyB, PhyA dimerizes through head-to-head assembly of its C-terminal histidine kinase-related domains (HKRDs), while the remainder assembles as a head-to-tail light-responsive platform. Whereas the platform and HKRDs associate asymmetrically in PhyB dimers, these lopsided connections are absent in PhyA. Analysis of truncation and site-directed mutants revealed that this decoupling and altered platform assembly have functional consequences for Pfr stability of PhyA and highlights how plant Phy structural diversification has extended light and temperature perception.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures







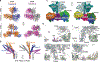




Similar articles
-
Plant phytochrome B is an asymmetric dimer with unique signalling potential.Nature. 2022 Apr;604(7904):127-133. doi: 10.1038/s41586-022-04529-z. Epub 2022 Mar 30. Nature. 2022. PMID: 35355010 Free PMC article.
-
Both phytochrome A and phyB interact with PHYTOCHROME-INTERACTING FACTORs through an evolutionary conserved phyOPM-APA interaction.Nat Commun. 2025 Apr 26;16(1):3946. doi: 10.1038/s41467-025-59327-8. Nat Commun. 2025. PMID: 40287465 Free PMC article.
-
Arabidopsis phytochrome a is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome.Plant Cell. 2012 Jul;24(7):2949-62. doi: 10.1105/tpc.111.094201. Epub 2012 Jul 27. Plant Cell. 2012. PMID: 22843485 Free PMC article.
-
The system of phytochromes: photobiophysics and photobiochemistry in vivo.Membr Cell Biol. 1998;12(5):691-720. Membr Cell Biol. 1998. PMID: 10379648 Review.
-
Two Distinct Molecular Types of Phytochrome A in Plants: Evidence of Existence and Implications for Functioning.Int J Mol Sci. 2023 May 2;24(9):8139. doi: 10.3390/ijms24098139. Int J Mol Sci. 2023. PMID: 37175844 Free PMC article. Review.
Cited by
-
Signaling by a bacterial phytochrome histidine kinase involves a conformational cascade reorganizing the dimeric photoreceptor.Nat Commun. 2024 Aug 10;15(1):6853. doi: 10.1038/s41467-024-50412-y. Nat Commun. 2024. PMID: 39127720 Free PMC article.
-
Light signaling in plants-a selective history.Plant Physiol. 2024 Apr 30;195(1):213-231. doi: 10.1093/plphys/kiae110. Plant Physiol. 2024. PMID: 38431282 Free PMC article. Review.
-
Structural insight into PIF6-mediated red light signal transduction of plant phytochrome B.Cell Discov. 2025 May 22;11(1):51. doi: 10.1038/s41421-025-00802-3. Cell Discov. 2025. PMID: 40404641 Free PMC article.
-
Plant Phytochrome Interactions Decode Light and Temperature Signals.Plant Cell. 2024 Sep 11;36(12):4819-39. doi: 10.1093/plcell/koae249. Online ahead of print. Plant Cell. 2024. PMID: 39259296 Free PMC article.
-
Review of protein structure-based analyses that illuminate plant stress mechanisms.Comput Struct Biotechnol J. 2025 Jul 13;27:3155-3166. doi: 10.1016/j.csbj.2025.07.021. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40727428 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

