A novel copper-induced cell death-related lncRNA prognostic signature associated with immune infiltration and clinical value in gastric cancer
- PMID: 37291405
- PMCID: PMC10423106
- DOI: 10.1007/s00432-023-04916-7
A novel copper-induced cell death-related lncRNA prognostic signature associated with immune infiltration and clinical value in gastric cancer
Abstract
Background: Gastric cancer (GC) is one of the most important malignancies and has a poor prognosis. Copper-induced cell death, recently termed cuproptosis, may directly affect the outcome of GC. Long noncoding RNAs (lncRNAs), possessing stable structures, can influence the prognosis of cancer and may serve as potential prognostic prediction factors for various cancers. However, the role of copper cell death-related lncRNAs (CRLs) in GC has not been thoroughly investigated. Here, we aim to elucidate the role of CRLs in predicting prognosis, diagnosis, and immunotherapy in GC patients.
Methods: RNA expression data for 407 GC patients from The Cancer Genome Atlas (TCGA) were gathered, and differentially expressed CRLs were identified. Subsequently, the researchers applied univariate, LASSO, and multivariate Cox regression to construct a prognostic signature consisting of 5 lncRNAs based on the CRLs. Stratified by the median CRLSig risk score, Kaplan-Meier analysis was utilized to compare overall survival (OS) between the high- and low-risk groups. Among the two groups, gene set enrichment analysis (GSEA), tumor microenvironment (TME), drug sensitivity analysis, and immune checkpoint analysis were conducted. In addition, consensus clustering and nomogram analysis were performed to predict OS. Cell experiments and 112 human serum samples were employed to verify the effect of lncRNAs on GC. Furthermore, the diagnostic value of the CRLSig in the serum of GC patients was analyzed by the receiver operating characteristic (ROC) curve.
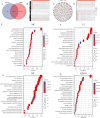
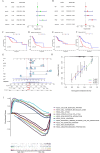
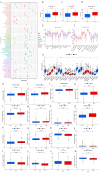
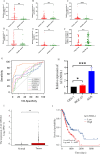
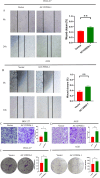
Results: A prognostic signature for GC patients was constructed based on CRLs, composed of AC129926.1, AP002954.1, AC023511.1, LINC01537, and TMEM75. According to the K-M survival analysis, high-risk GC patients had a lower OS rate and progression-free survival rate than low-risk GC patients. Further support for the model's accuracy was provided by ROC, principal component analysis, and the validation set. The area under the curve (AUC) of 0.772 for GC patients showed a better prognostic value than any other clinicopathological variable. Furthermore, immune infiltration analysis showed that the high-risk group had greater antitumor immune responses in the tumor microenvironment. In the high-risk subgroup, 23 immune checkpoint genes had significantly higher expression levels than in the low-risk subgroup (p < 0.05). The half-maximal inhibitory concentrations (IC50) of 86 drugs were found to be significantly different in the two groups. Accordingly, the model is capable of predicting the effectiveness of immunotherapy. In addition, the five CRLs in GC serum exhibited statistically significant expression levels. The AUC of this signature in GC serum was 0.894, with a 95% CI of 0.822-0.944. Moreover, lncRNA AC129926.1 was significantly overexpressed in GC cell lines and the serum of GC patients. Importantly, colony formation, wound healing, and transwell assays further confirmed the oncogenic role of AC129926.1 in GC.
Conclusion: In this study, a prognostic signature model consisting of five CRLs was developed to improve OS prediction accuracy in GC patients. The model also has the potential to predict immune infiltration and immunotherapy effectiveness. Furthermore, the CRLSig might serve as a novel serum biomarker to differentiate GC patients from healthy individuals.
Keywords: Biomarker; Copper induced cell death; Gastric cancer; Prognostic signature; lncRNA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures

References
-
- Ajani JA et al (2006) A multi-center phase II study of sequential paclitaxel and bryostatin-1 (NSC 339555) in patients with untreated, advanced gastric or gastroesophageal junction adenocarcinoma. Invest New Drugs 24(4):353–357 - PubMed
-
- Bach DH, Lee SK (2018) Long noncoding RNAs in cancer cells. Cancer Lett 419:152–166 - PubMed
-
- Beermann J et al (2016) Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 96(4):1297–1325 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

