Targeted and whole-genome sequencing reveal a north-south divide in P. falciparum drug resistance markers and genetic structure in Mozambique
- PMID: 37291425
- PMCID: PMC10250372
- DOI: 10.1038/s42003-023-04997-7
Targeted and whole-genome sequencing reveal a north-south divide in P. falciparum drug resistance markers and genetic structure in Mozambique
Abstract
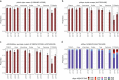
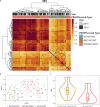
Mozambique is one of the four African countries which account for over half of all malaria deaths worldwide, yet little is known about the parasite genetic structure in that country. We performed P. falciparum amplicon and whole genome sequencing on 2251 malaria-infected blood samples collected in 2015 and 2018 in seven provinces of Mozambique to genotype antimalarial resistance markers and interrogate parasite population structure using genome-wide microhaplotyes. Here we show that the only resistance-associated markers observed at frequencies above 5% were pfmdr1-184F (59%), pfdhfr-51I/59 R/108 N (99%) and pfdhps-437G/540E (89%). The frequency of pfdhfr/pfdhps quintuple mutants associated with sulfadoxine-pyrimethamine resistance increased from 80% in 2015 to 89% in 2018 (p < 0.001), with a lower expected heterozygosity and higher relatedness of microhaplotypes surrounding pfdhps mutants than wild-type parasites suggestive of recent selection. pfdhfr/pfdhps quintuple mutants also increased from 72% in the north to 95% in the south (2018; p < 0.001). This resistance gradient was accompanied by a concentration of mutations at pfdhps-436 (17%) in the north, a south-to-north increase in the genetic complexity of P. falciparum infections (p = 0.001) and a microhaplotype signature of regional differentiation. The parasite population structure identified here offers insights to guide antimalarial interventions and epidemiological surveys.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- World Health Organization. WHO Malaria Report 2021 (WHO, 2022).
-
- World Health Organization. WHO Guidelines for malaria. WHO/UCN/GMP/2022.01 Rev.2 (2022).

