Protein length distribution is remarkably uniform across the tree of life
- PMID: 37291671
- PMCID: PMC10251718
- DOI: 10.1186/s13059-023-02973-2
Protein length distribution is remarkably uniform across the tree of life
Abstract
Background: In every living species, the function of a protein depends on its organization of structural domains, and the length of a protein is a direct reflection of this. Because every species evolved under different evolutionary pressures, the protein length distribution, much like other genomic features, is expected to vary across species but has so far been scarcely studied.
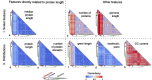
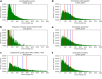
Results: Here we evaluate this diversity by comparing protein length distribution across 2326 species (1688 bacteria, 153 archaea, and 485 eukaryotes). We find that proteins tend to be on average slightly longer in eukaryotes than in bacteria or archaea, but that the variation of length distribution across species is low, especially compared to the variation of other genomic features (genome size, number of proteins, gene length, GC content, isoelectric points of proteins). Moreover, most cases of atypical protein length distribution appear to be due to artifactual gene annotation, suggesting the actual variation of protein length distribution across species is even smaller.
Conclusions: These results open the way for developing a genome annotation quality metric based on protein length distribution to complement conventional quality measures. Overall, our findings show that protein length distribution between living species is more uniform than previously thought. Furthermore, we also provide evidence for a universal selection on protein length, yet its mechanism and fitness effect remain intriguing open questions.
Keywords: Comparative genomics; Genome annotation; Genome evolution; Protein length.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Wright SI. Evolution of Genome Size [Internet]. eLS. Chichester, UK: John Wiley & Sons, Ltd; 2017. p. 1–6. Available from: 10.1002/9780470015902.a0023983
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

