Genome sequencing of Enterococcus faecium NT04, an oral microbiota revealed the production of enterocin A/B active against oral pathogens
- PMID: 37292348
- PMCID: PMC10245172
- DOI: 10.1016/j.heliyon.2023.e16253
Genome sequencing of Enterococcus faecium NT04, an oral microbiota revealed the production of enterocin A/B active against oral pathogens
Abstract
Objective: This study aimed to isolate and investigate a bacterium from an Egyptian adult's healthy oral cavity, focusing on its probiotic properties, especially its antagonistic activity against oral pathogens.
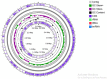
Methods: The isolated bacterium NT04 using 16S rRNA gene sequencing, was identified as Enterococcus faecium. In this study, the whole genome of Enterococcus faecium NT04 was sequenced and annotated by bioinformatics analysis tools.
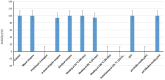
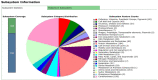
Results: Numerous genes encoding the production of diverse metabolic and probiotic properties, such as bacteriocin-like inhibitory substances (Enterocin A and B), cofactors, antioxidants, and vitamins, were confirmed by genomic analysis. There were no pathogenicity islands or plasmid insertions found. This strain is virulent for host colonization rather than invasion.
Conclusion: Genomic characteristics of strain NT04 support its potentiality as an anti-oral pathogen probiotic candidate.
Keywords: Enterococcus faecium; Oral cavity; Probiotics; WGS.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Baran A., et al. Proteolytic activities and safety use of Enterococcus faecalis strains isolated from Turkish white pickled cheese and milk samples. J. Hellenic Veter. Med. Soc. 2022;72(4):3391–3400.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

