This is a preprint.
Microsecond dynamics control the HIV-1 envelope conformation
- PMID: 37292605
- PMCID: PMC10245784
- DOI: 10.1101/2023.05.17.541130
Microsecond dynamics control the HIV-1 envelope conformation
Update in
-
Microsecond dynamics control the HIV-1 Envelope conformation.Sci Adv. 2024 Feb 2;10(5):eadj0396. doi: 10.1126/sciadv.adj0396. Epub 2024 Feb 2. Sci Adv. 2024. PMID: 38306419 Free PMC article.
Abstract
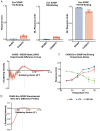
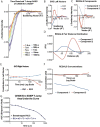
The HIV-1 Envelope (Env) glycoprotein facilitates host cell fusion through a complex series of receptor-induced structural changes. Although significant progress has been made in understanding the structures of various Env conformations and transition intermediates that occur within the millisecond timescale, faster transitions in the microsecond timescale have not yet been observed. In this study, we employed time-resolved, temperature-jump small angle X-ray scattering to monitor structural rearrangements in an HIV-1 Env ectodomain construct with microsecond precision. We detected a transition correlated with Env opening that occurs in the hundreds of microseconds range and another more rapid transition that preceded this opening. Model fitting indicated that the early rapid transition involved an order-to-disorder transition in the trimer apex loop contacts, suggesting that conventional conformation-locking design strategies that target the allosteric machinery may be ineffective in preventing this movement. Utilizing this information, we engineered an envelope that locks the apex loop contacts to the adjacent protomer. This modification resulted in significant angle-of-approach shifts in the interaction of a neutralizing antibody. Our findings imply that blocking the intermediate state could be crucial for inducing antibodies with the appropriate bound state orientation through vaccination.
Conflict of interest statement
Competing Interest The authors declare the following competing interests: A patent application covering HIV-1 Envelope modifications based on this study has been submitted by Duke University.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
