This is a preprint.
Integrated multimodal cell atlas of Alzheimer's disease
- PMID: 37292694
- PMCID: PMC10246227
- DOI: 10.21203/rs.3.rs-2921860/v1
Integrated multimodal cell atlas of Alzheimer's disease
Update in
-
Integrated multimodal cell atlas of Alzheimer's disease.Nat Neurosci. 2024 Dec;27(12):2366-2383. doi: 10.1038/s41593-024-01774-5. Epub 2024 Oct 14. Nat Neurosci. 2024. PMID: 39402379 Free PMC article.
Abstract
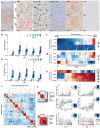
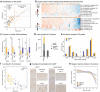
Alzheimer's disease (AD) is the most common cause of dementia in older adults. Neuropathological and imaging studies have demonstrated a progressive and stereotyped accumulation of protein aggregates, but the underlying molecular and cellular mechanisms driving AD progression and vulnerable cell populations affected by disease remain coarsely understood. The current study harnesses single cell and spatial genomics tools and knowledge from the BRAIN Initiative Cell Census Network to understand the impact of disease progression on middle temporal gyrus cell types. We used image-based quantitative neuropathology to place 84 donors spanning the spectrum of AD pathology along a continuous disease pseudoprogression score and multiomic technologies to profile single nuclei from each donor, mapping their transcriptomes, epigenomes, and spatial coordinates to a common cell type reference with unprecedented resolution. Temporal analysis of cell-type proportions indicated an early reduction of Somatostatin-expressing neuronal subtypes and a late decrease of supragranular intratelencephalic-projecting excitatory and Parvalbumin-expressing neurons, with increases in disease-associated microglial and astrocytic states. We found complex gene expression differences, ranging from global to cell type-specific effects. These effects showed different temporal patterns indicating diverse cellular perturbations as a function of disease progression. A subset of donors showed a particularly severe cellular and molecular phenotype, which correlated with steeper cognitive decline. We have created a freely available public resource to explore these data and to accelerate progress in AD research at SEA-AD.org.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures



References
-
- Masters C. L. et al. Alzheimer’s disease. Nature Reviews Disease Primers 1, 15056 (2015). - PubMed
-
- Thal D. R., Rub U., Orantes M. & Braak H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology 58, 1791–1800 (2002). - PubMed
-
- Braak H. & Braak E. Neuropathological stageing of Alzheimer-related changes. Acta neuropathologica 82, 239–259 (1991). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

