This is a preprint.
Within-host SARS-CoV-2 viral kinetics informed by complex life course exposures reveals different intrinsic properties of Omicron and Delta variants
- PMID: 37292842
- PMCID: PMC10246130
- DOI: 10.1101/2023.05.17.23290105
Within-host SARS-CoV-2 viral kinetics informed by complex life course exposures reveals different intrinsic properties of Omicron and Delta variants
Abstract
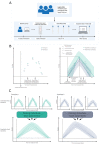
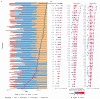
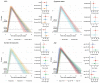
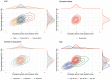
The emergence of successive SARS-CoV-2 variants of concern (VOC) during 2020-22, each exhibiting increased epidemic growth relative to earlier circulating variants, has created a need to understand the drivers of such growth. However, both pathogen biology and changing host characteristics - such as varying levels of immunity - can combine to influence replication and transmission of SARS-CoV-2 within and between hosts. Disentangling the role of variant and host in individual-level viral shedding of VOCs is essential to inform COVID-19 planning and response, and interpret past epidemic trends. Using data from a prospective observational cohort study of healthy adult volunteers undergoing weekly occupational health PCR screening, we developed a Bayesian hierarchical model to reconstruct individual-level viral kinetics and estimate how different factors shaped viral dynamics, measured by PCR cycle threshold (Ct) values over time. Jointly accounting for both inter-individual variation in Ct values and complex host characteristics - such as vaccination status, exposure history and age - we found that age and number of prior exposures had a strong influence on peak viral replication. Older individuals and those who had at least five prior antigen exposures to vaccination and/or infection typically had much lower levels of shedding. Moreover, we found evidence of a correlation between the speed of early shedding and duration of incubation period when comparing different VOCs and age groups. Our findings illustrate the value of linking information on participant characteristics, symptom profile and infecting variant with prospective PCR sampling, and the importance of accounting for increasingly complex population exposure landscapes when analysing the viral kinetics of VOCs.
Figures
References
-
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021. May;593(7858):266–9. - PubMed
-
- Kucharski AJ, Davies NG, Eggo RM, Funk S, CMMID working group Dynamics of B.1.617.2 in the UK from importations, traveller-linked and non-traveller-linked transmission [Internet]. London School of Hygiene and Tropical Medicine; 2021. May. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploa...
-
- Killingley B, Mann AJ, Kalinova M, Boyers A, Goonawardane N, Zhou J, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. 2022. May;28(5):1031–41. - PubMed
-
- Hay JA, Kissler SM, Fauver JR, Mack C, Tai CG, Samant RM, et al. Viral dynamics and duration of PCR positivity of the SARS-CoV-2 Omicron variant [Internet]. Epidemiology; 2022. Jan [cited 2022 Jun 20]. Available from: 10.1101/2022.01.13.22269257 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
