This is a preprint.
Extended Nucleic Acid (exNA): A Novel, Biologically Compatible Backbone that Significantly Enhances Oligonucleotide Efficacy in vivo
- PMID: 37292886
- PMCID: PMC10245983
- DOI: 10.1101/2023.05.26.542506
Extended Nucleic Acid (exNA): A Novel, Biologically Compatible Backbone that Significantly Enhances Oligonucleotide Efficacy in vivo
Update in
-
Enhancing siRNA efficacy in vivo with extended nucleic acid backbones.Nat Biotechnol. 2025 Jun;43(6):904-913. doi: 10.1038/s41587-024-02336-7. Epub 2024 Aug 1. Nat Biotechnol. 2025. PMID: 39090305
Abstract
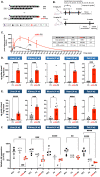
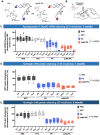
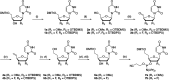
Metabolic stabilization of therapeutic oligonucleotides requires both sugar and backbone modifications, where phosphorothioate (PS) is the only backbone chemistry used in the clinic. Here, we describe the discovery, synthesis, and characterization of a novel biologically compatible backbone, extended nucleic acid (exNA). Upon exNA precursor scale up, exNA incorporation is fully compatible with common nucleic acid synthetic protocols. The novel backbone is orthogonal to PS and shows profound stabilization against 3'- and 5'-exonucleases. Using small interfering RNAs (siRNAs) as an example, we show exNA is tolerated at most nucleotide positions and profoundly improves in vivo efficacy. A combined exNA-PS backbone enhances siRNA resistance to serum 3'-exonuclease by ~32-fold over PS backbone and >1000-fold over the natural phosphodiester backbone, thereby enhancing tissue exposure (~6-fold), tissues accumulation (4- to 20-fold), and potency both systemically and in brain. The improved potency and durability imparted by exNA opens more tissues and indications to oligonucleotide-driven therapeutic interventions.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
