This is a preprint.
Development of a genetically encoded sensor for probing endogenous nociceptin opioid peptide release
- PMID: 37292957
- PMCID: PMC10245933
- DOI: 10.1101/2023.05.26.542102
Development of a genetically encoded sensor for probing endogenous nociceptin opioid peptide release
Update in
-
Development of a genetically encoded sensor for probing endogenous nociceptin opioid peptide release.Nat Commun. 2024 Jun 25;15(1):5353. doi: 10.1038/s41467-024-49712-0. Nat Commun. 2024. PMID: 38918403 Free PMC article.
Abstract
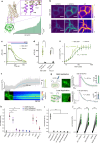
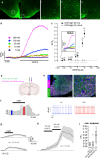
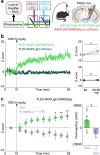
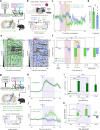
Nociceptin/orphanin-FQ (N/OFQ) is a recently appreciated critical opioid peptide with key regulatory functions in several central behavioral processes including motivation, stress, feeding, and sleep. The functional relevance of N/OFQ action in the mammalian brain remains unclear due to a lack of high-resolution approaches to detect this neuropeptide with appropriate spatial and temporal resolution. Here we develop and characterize NOPLight, a genetically encoded sensor that sensitively reports changes in endogenous N/OFQ release. We characterized the affinity, pharmacological profile, spectral properties, kinetics, ligand selectivity, and potential interaction with intracellular signal transducers of NOPLight in vitro. Its functionality was established in acute brain slices by exogeneous N/OFQ application and chemogenetic induction of endogenous N/OFQ release from PNOC neurons. In vivo studies with fibre photometry enabled direct recording of NOPLight binding to exogenous N/OFQ receptor ligands, as well as detection of endogenous N/OFQ release within the paranigral ventral tegmental area (pnVTA) during natural behaviors and chemogenetic activation of PNOC neurons. In summary, we show here that NOPLight can be used to detect N/OFQ opioid peptide signal dynamics in tissue and freely behaving animals.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
