This is a preprint.
VirB, a key transcriptional regulator of Shigella virulence, requires a CTP ligand for its regulatory activities
- PMID: 37293012
- PMCID: PMC10245682
- DOI: 10.1101/2023.05.16.541010
VirB, a key transcriptional regulator of Shigella virulence, requires a CTP ligand for its regulatory activities
Update in
-
VirB, a key transcriptional regulator of Shigella virulence, requires a CTP ligand for its regulatory activities.mBio. 2023 Oct 31;14(5):e0151923. doi: 10.1128/mbio.01519-23. Epub 2023 Sep 20. mBio. 2023. PMID: 37728345 Free PMC article.
Abstract
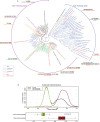
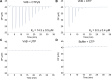
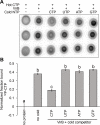
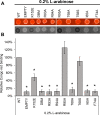
The VirB protein, encoded by the large virulence plasmid of Shigella spp., is a key transcriptional regulator of virulence genes. Without a functional virB gene, Shigella cells are avirulent. On the virulence plasmid, VirB functions to offset transcriptional silencing mediated by the nucleoid structuring protein, H-NS, which binds and sequesters AT-rich DNA, making it inaccessible for gene expression. Thus, gaining a mechanistic understanding of how VirB counters H-NS-mediated silencing is of considerable interest. VirB is unusual in that it does not resemble classic transcription factors. Instead, its closest relatives are found in the ParB superfamily, where the best-characterized members function in faithful DNA segregation before cell division. Here, we show that VirB is a fast-evolving member of this superfamily and report for the first time that the VirB protein binds a highly unusual ligand, CTP. VirB binds this nucleoside triphosphate preferentially and with specificity. Based on alignments with the best-characterized members of the ParB family, we identify amino acids of VirB likely to bind CTP. Substitutions in these residues disrupt several well-documented activities of VirB, including its anti-silencing activity at a VirB-dependent promoter, its role in generating a Congo red positive phenotype in Shigella , and the ability of the VirB protein to form foci in the bacterial cytoplasm when fused to GFP. Thus, this work is the first to show that VirB is a bona fide CTP-binding protein and links Shigella virulence phenotypes to the nucleoside triphosphate, CTP.
Importance: Shigella species cause bacillary dysentery (shigellosis), the second leading cause of diarrheal deaths worldwide. With growing antibiotic resistance, there is a pressing need to identify novel molecular drug targets. Shigella virulence phenotypes are controlled by the transcriptional regulator, VirB. We show that VirB belongs to a fast-evolving, primarily plasmid-borne clade of the ParB superfamily, which has diverged from versions that have a distinct cellular role - DNA partitioning. We are the first to report that, like classic members of the ParB family, VirB binds a highly unusual ligand, CTP. Mutants predicted to be defective in CTP binding are compromised in a variety of virulence attributes controlled by VirB. This study i) reveals that VirB binds CTP, ii) provides a link between VirB-CTP interactions and Shigella virulence phenotypes, and iii) broadens our understanding of the ParB superfamily, a group of bacterial proteins that play critical roles in many different bacteria.
Figures


References
-
- Niyogi SK. 2007. Increasing antimicrobial resistance--an emerging problem in the treatment of shigellosis. Clin Microbiol Infect 13:1141–3. - PubMed
-
- Niyogi SK. 2005. Shigellosis. J Microbiol 43:133–43. - PubMed
-
- Tobe T, Nagai S, Okada N, Adler B, Yoshikawa M, Sasakawa C. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol Microbiol 5:887–93. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
