This is a preprint.
PfAP2-MRP DNA-binding protein is a master regulator of parasite pathogenesis during malaria parasite blood stages
- PMID: 37293082
- PMCID: PMC10245809
- DOI: 10.1101/2023.05.23.541898
PfAP2-MRP DNA-binding protein is a master regulator of parasite pathogenesis during malaria parasite blood stages
Update in
-
DNA-binding protein PfAP2-P regulates parasite pathogenesis during malaria parasite blood stages.Nat Microbiol. 2023 Nov;8(11):2154-2169. doi: 10.1038/s41564-023-01497-6. Epub 2023 Oct 26. Nat Microbiol. 2023. PMID: 37884813 Free PMC article.
Abstract
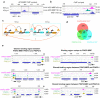
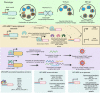
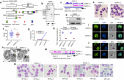
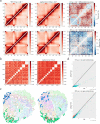
Malaria pathogenicity results from the parasite's ability to invade, multiply within and then egress from the host red blood cell (RBC). Infected RBCs are remodeled, expressing antigenic variant proteins (such as PfEMP1, coded by the var gene family) for immune evasion and survival. These processes require the concerted actions of many proteins, but the molecular regulation is poorly understood. We have characterized an essential Plasmodium specific Apicomplexan AP2 (ApiAP2) transcription factor in Plasmodium falciparum (PfAP2-MRP; Master Regulator of Pathogenesis) during the intraerythrocytic developmental cycle (IDC). An inducible gene knockout approach showed that PfAP2-MRP is essential for development during the trophozoite stage, and critical for var gene regulation, merozoite development and parasite egress. ChIP-seq experiments performed at 16 hour post invasion (h.p.i.) and 40 h.p.i. matching the two peaks of PfAP2-MRP expression, demonstrate binding of PfAP2-MRP to the promoters of genes controlling trophozoite development and host cell remodeling at 16 h.p.i. and antigenic variation and pathogenicity at 40 h.p.i. Using single-cell RNA-seq and fluorescence-activated cell sorting, we show de-repression of most var genes in Δpfap2-mrp parasites that express multiple PfEMP1 proteins on the surface of infected RBCs. In addition, the Δpfap2-mrp parasites overexpress several early gametocyte marker genes at both 16 and 40 h.p.i., indicating a regulatory role in the sexual stage conversion. Using the Chromosomes Conformation Capture experiment (Hi-C), we demonstrate that deletion of PfAP2-MRP results in significant reduction of both intra-chromosomal and inter-chromosomal interactions in heterochromatin clusters. We conclude that PfAP2-MRP is a vital upstream transcriptional regulator controlling essential processes in two distinct developmental stages during the IDC that include parasite growth, chromatin structure and var gene expression.
Figures

















References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
