Ubiquitin E3 ligase activity of Ralstonia solanacearum effector RipAW is not essential for induction of plant defense in Nicotiana benthamiana
- PMID: 37293211
- PMCID: PMC10244751
- DOI: 10.3389/fmicb.2023.1201444
Ubiquitin E3 ligase activity of Ralstonia solanacearum effector RipAW is not essential for induction of plant defense in Nicotiana benthamiana
Abstract
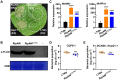
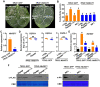
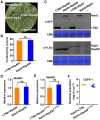
As one of the most destructive bacterial phytopathogens, Ralstonia solanacearum causes substantial annual yield losses of many important crops. Deciphering the functional mechanisms of type III effectors, the crucial factors mediating R. solanacearum-plant interactions, will provide a valuable basis for protecting crop plants from R. solanacearum. Recently, the NEL (novel E3 ligase) effector RipAW was found to induce cell death on Nicotiana benthamiana in a E3 ligase activity-dependent manner. Here, we further deciphered the role of the E3 ligase activity in RipAW-triggered plant immunity. We found that RipAWC177A, the E3 ligase mutant of RipAW, could not induce cell death but retained the ability of triggering plant immunity in N. benthamiana, indicating that the E3 ligase activity is not essential for RipAW-triggered immunity. By generating truncated mutants of RipAW, we further showed that the N-terminus, NEL domain and C-terminus are all required but not sufficient for RipAW-induced cell death. Furthermore, all truncated mutants of RipAW triggered ETI immune responses in N. benthamiana, confirming that the E3 ligase activity is not essential for RipAW-triggered plant immunity. Finally, we demonstrated that RipAW- and RipAWC177A-triggered immunity in N. benthamiana requires SGT1 (suppressor of G2 allele of skp1), but not EDS1 (enhanced disease susceptibility), NRG1 (N requirement gene 1), NRC (NLR required for cell death) proteins or SA (salicylic acid) pathway. Our findings provide a typical case in which the effector-induced cell death can be uncoupled with immune responses, shedding new light on effector-triggered plant immunity. Our data also provide clues for further in-depth study of mechanism underlying RipAW-induced plant immunity.
Keywords: Ralstonia solanacearum; RipAW; SGT1; cell death; plant immunity.
Copyright © 2023 Ouyang, Chen, Sun, Wang, Wu, Li, Song, Liu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
The Ralstonia solanacearum Type III Effector RipAW Targets the Immune Receptor Complex to Suppress PAMP-Triggered Immunity.Int J Mol Sci. 2023 Dec 22;25(1):183. doi: 10.3390/ijms25010183. Int J Mol Sci. 2023. PMID: 38203354 Free PMC article.
-
Different epitopes of Ralstonia solanacearum effector RipAW are recognized by two Nicotiana species and trigger immune responses.Mol Plant Pathol. 2022 Feb;23(2):188-203. doi: 10.1111/mpp.13153. Epub 2021 Oct 31. Mol Plant Pathol. 2022. PMID: 34719088 Free PMC article.
-
Ralstonia solanacearum novel E3 ubiquitin ligase (NEL) effectors RipAW and RipAR suppress pattern-triggered immunity in plants.Microbiology (Reading). 2017 Jul;163(7):992-1002. doi: 10.1099/mic.0.000495. Epub 2017 Jul 21. Microbiology (Reading). 2017. PMID: 28708051
-
Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era.Mol Plant Pathol. 2013 Sep;14(7):651-62. doi: 10.1111/mpp.12038. Epub 2013 May 30. Mol Plant Pathol. 2013. PMID: 23718203 Free PMC article. Review.
-
Breeding for resistances to Ralstonia solanacearum.Front Plant Sci. 2014 Dec 12;5:715. doi: 10.3389/fpls.2014.00715. eCollection 2014. Front Plant Sci. 2014. PMID: 25566289 Free PMC article. Review.
Cited by
-
The Ralstonia solanacearum Type III Effector RipAW Targets the Immune Receptor Complex to Suppress PAMP-Triggered Immunity.Int J Mol Sci. 2023 Dec 22;25(1):183. doi: 10.3390/ijms25010183. Int J Mol Sci. 2023. PMID: 38203354 Free PMC article.
-
Molecular Dialog of Ralstonia solanacearum and Plant Hosts with Highlights on Type III Effectors.Int J Mol Sci. 2025 Apr 13;26(8):3686. doi: 10.3390/ijms26083686. Int J Mol Sci. 2025. PMID: 40332227 Free PMC article. Review.
-
Mechanism by which SUGT1 downregulates FH to promote proliferation and migration in serous ovarian cancer.J Ovarian Res. 2025 Jul 29;18(1):168. doi: 10.1186/s13048-025-01744-w. J Ovarian Res. 2025. PMID: 40731365 Free PMC article.
References
-
- Bos J. I. B., Kanneganti T. D., Young C., Cakir C., Huitema E., Win J., et al. . (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48, 165–176. doi: 10.1111/j.1365-313X.2006.02866.x, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

