Evidence of horizontal gene transfer within porB in 19 018 whole-genome Neisseria spp. isolates: a global phylogenetic analysis
- PMID: 37294009
- PMCID: PMC10327514
- DOI: 10.1099/mgen.0.001041
Evidence of horizontal gene transfer within porB in 19 018 whole-genome Neisseria spp. isolates: a global phylogenetic analysis
Abstract
The PorB porins are the major pore-forming proteins in the genus
Keywords: Neisseria; PorB1a; Porb1b; commensal Neisseria; phylogenetics; porin.
Conflict of interest statement
The author(s) declare that there are no conflicts of interest.
Figures
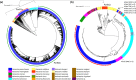



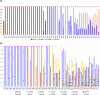
Similar articles
-
Clusters of circulating Neisseria gonorrhoeae strains and association with antimicrobial resistance in Shanghai.J Antimicrob Chemother. 2008 Mar;61(3):478-87. doi: 10.1093/jac/dkm544. Epub 2008 Jan 27. J Antimicrob Chemother. 2008. PMID: 18227091
-
Molecular typing of Neisseria gonorrhoeae isolates by pyrosequencing of highly polymorphic segments of the porB gene.J Clin Microbiol. 2004 Jul;42(7):2926-34. doi: 10.1128/JCM.42.7.2926-2934.2004. J Clin Microbiol. 2004. PMID: 15243040 Free PMC article.
-
Comprehensive Bioinformatic Assessments of the Variability of Neisseria gonorrhoeae Vaccine Candidates.mSphere. 2021 Feb 3;6(1):e00977-20. doi: 10.1128/mSphere.00977-20. mSphere. 2021. PMID: 33536323 Free PMC article.
-
Canary in the Coal Mine: How Resistance Surveillance in Commensals Could Help Curb the Spread of AMR in Pathogenic Neisseria.mBio. 2022 Oct 26;13(5):e0199122. doi: 10.1128/mbio.01991-22. Epub 2022 Sep 26. mBio. 2022. PMID: 36154280 Free PMC article. Review.
-
Mobile genetic elements in Neisseria gonorrhoeae: movement for change.Pathog Dis. 2017 Aug 31;75(6). doi: 10.1093/femspd/ftx071. Pathog Dis. 2017. PMID: 28645177 Review.
Cited by
-
Genome-wide investigation of outer membrane protein families under mosaic evolution in Escherichia coli.Appl Environ Microbiol. 2025 Jun 18;91(6):e0055725. doi: 10.1128/aem.00557-25. Epub 2025 May 30. Appl Environ Microbiol. 2025. PMID: 40444982 Free PMC article.
-
Non-pathogenic Neisseria species of the oropharynx as a reservoir of antimicrobial resistance: a cross-sectional study.Front Cell Infect Microbiol. 2023 Nov 22;13:1308550. doi: 10.3389/fcimb.2023.1308550. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38076458 Free PMC article.
-
A Meta-analysis to Quantify the Risk of Disseminated Gonococcal Infection With Porin B Serotype.Open Forum Infect Dis. 2024 Jul 8;11(7):ofae389. doi: 10.1093/ofid/ofae389. eCollection 2024 Jul. Open Forum Infect Dis. 2024. PMID: 39035573 Free PMC article.
-
Leveraging the microbiome to combat antibiotic resistant gynecological infections.NPJ Antimicrob Resist. 2025 Apr 23;3(1):32. doi: 10.1038/s44259-025-00106-2. NPJ Antimicrob Resist. 2025. PMID: 40269132 Free PMC article. Review.
-
Gonococcal PorB: a multifaceted modulator of host immune responses.Trends Microbiol. 2024 Apr;32(4):355-364. doi: 10.1016/j.tim.2023.10.002. Epub 2023 Oct 25. Trends Microbiol. 2024. PMID: 37891023 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

