The pros and cons of ubiquitination on the formation of protein condensates
- PMID: 37294105
- PMCID: PMC10423694
- DOI: 10.3724/abbs.2023096
The pros and cons of ubiquitination on the formation of protein condensates
Abstract
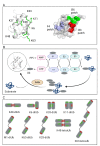
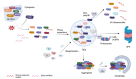
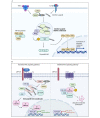
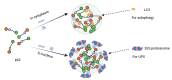
Ubiquitination, a post-translational modification that attaches one or more ubiquitin (Ub) molecules to another protein, plays a crucial role in the phase-separation processes. Ubiquitination can modulate the formation of membrane-less organelles in two ways. First, a scaffold protein drives phase separation, and Ub is recruited to the condensates. Second, Ub actively phase-separates through the interactions with other proteins. Thus, the role of ubiquitination and the resulting polyUb chains ranges from bystanders to active participants in phase separation. Moreover, long polyUb chains may be the primary driving force for phase separation. We further discuss that the different roles can be determined by the lengths and linkages of polyUb chains which provide preorganized and multivalent binding platforms for other client proteins. Together, ubiquitination adds a new layer of regulation for the flow of material and information upon cellular compartmentalization of proteins.
Keywords: phase separation; polyubiquitin chain; post-translational modification; stress granule; ubiquitination.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units.Proc Natl Acad Sci U S A. 2023 Oct 17;120(42):e2306638120. doi: 10.1073/pnas.2306638120. Epub 2023 Oct 12. Proc Natl Acad Sci U S A. 2023. PMID: 37824531 Free PMC article.
-
Analysis of the biochemical role of Lys-11 in polyubiquitin chain formation using quantitative mass spectrometry.Rapid Commun Mass Spectrom. 2013 Jan 30;27(2):339-46. doi: 10.1002/rcm.6447. Rapid Commun Mass Spectrom. 2013. PMID: 23239382
-
Decoding optimal ligand design for multicomponent condensates.bioRxiv [Preprint]. 2023 Apr 25:2023.03.13.532222. doi: 10.1101/2023.03.13.532222. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Oct 17;120(42):e2306638120. doi: 10.1073/pnas.2306638120. PMID: 36993708 Free PMC article. Updated. Preprint.
-
Proteasome-independent functions of lysine-63 polyubiquitination in plants.New Phytol. 2018 Feb;217(3):995-1011. doi: 10.1111/nph.14915. Epub 2017 Nov 30. New Phytol. 2018. PMID: 29194634 Review.
-
The missing links to link ubiquitin: Methods for the enzymatic production of polyubiquitin chains.Anal Biochem. 2016 Jan 1;492:82-90. doi: 10.1016/j.ab.2015.09.013. Epub 2015 Oct 20. Anal Biochem. 2016. PMID: 26470940 Review.
Cited by
-
Liquid-Liquid Phase Separation Sheds New Light upon Cardiovascular Diseases.Int J Mol Sci. 2023 Oct 21;24(20):15418. doi: 10.3390/ijms242015418. Int J Mol Sci. 2023. PMID: 37895097 Free PMC article. Review.
-
Phase separation promotes the activity of HECT E3 ligases-A word of caution.Proc Natl Acad Sci U S A. 2024 Apr 2;121(14):e2402551121. doi: 10.1073/pnas.2402551121. Epub 2024 Mar 25. Proc Natl Acad Sci U S A. 2024. PMID: 38527205 Free PMC article. No abstract available.
-
The riverdance of proteins within the cell: a special issue on protein phase separation in biology and diseases.Acta Biochim Biophys Sin (Shanghai). 2023 Jul 25;55(7):1021-1022. doi: 10.3724/abbs.2023140. Acta Biochim Biophys Sin (Shanghai). 2023. PMID: 37489010 Free PMC article. No abstract available.
-
Caspase-2 is a condensate-mediated deubiquitinase in protein quality control.Nat Cell Biol. 2024 Nov;26(11):1943-1957. doi: 10.1038/s41556-024-01522-8. Epub 2024 Oct 31. Nat Cell Biol. 2024. PMID: 39482354 Free PMC article.
References
-
- Tong X, Tang R, Xu J, Wang W, Zhao Y, Yu X, Shi S. Liquid–liquid phase separation in tumor biology. Sig Transduct Target Ther. . 2022;7:221. doi: 10.1038/s41392-022-01076-x. - DOI - PMC - PubMed
-
- Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. . 2017;18:285–298. doi: 10.1038/nrm.2017.7. - DOI - PMC - PubMed
-
- Deliz-Aguirre R, Cao F, Gerpott FHU, Auevechanichkul N, Chupanova M, Mun YV, Ziska E, et al. MyD88 oligomer size functions as a physical threshold to trigger IL1R Myddosome signaling. J Cell Biol. . 2021;220:e202012071. doi: 10.1083/jcb.202012071. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

